`summarise()` has grouped output by 'date'. You can override using the
`.groups` argument.Exercises Day 5
Notes and instructions
It is expected that the exercise answers will be mainly using the pipe operators |>, instead of putting functions inside each other
Dataset
We will use covid_cases.rds for this exercise.
Data dictionary
| Column | Description |
|---|---|
date |
date of the case report |
211 columns with the format cases_{country_code} |
number of reported cases of each country and archipelago |
Country codes of interest (i.e. Codes for countries needed for the exercise)
| Code | Country name |
|---|---|
chn |
China |
usa |
United States of America |
vnm |
Vietnam |
sgp |
Singapore |
Exercises
Task 1: Data import
Read data
Load the
tidyversemeta-package into R
A “meta-package” is a package that contains other packages. Remember, tidyverse is just a collection of packages.
Task 2: Data cleaning and filtering
Does the data follow the tidy data standard?
- If not, pivot the data into a
tibblethat follows the tidy data standard
- If not, pivot the data into a
Do some quick skim on the data. Do the variable values make sense given its type?
- If not, filter out incorrect/impossible data
Filter the data so that we only have week 3-12 of 2020
Save the results back into
covid_cases
If your data is not tidy data:
“Lengthens” the data using
tidyr::pivot_longer(), i.e. less columns, more rows“Widens” the data using
tidyr::pivot_wider(), i.e. less rows, more columns
The main variable here is case counts from each country. They should have a
numerictype and should not be lower than 0. You can’t have negative case countsYou can remove or filter undesirable values using
dplyr::filter()
Task 3: Data transformation
Using the
covid_casesobject created in Task 2:Group the data and calculate the total number of cases per country
Select the top 5 countries with highest total cases
Extract the country codes and save them into a new object, e.g.
top_countries
Use the
covid_casesobject created in Task 2 again:Group the data again, calculate the total number of cases per country again
Modify the
countrycolumn:So that we only keep the names of countries in the top 5 (generated above). Countries on in the top 5 will be changed to
"Others"Turn this column into a
factortype using theforcatspackage (already intidyverse)Group the data again, this time, group by date and country
Calculate the total number of cases per date per country
Create a new column called
pct_cases, which is the percentage of total cases per date per country(Optional) remove rows with NA from the
tibbleThere are many ways to do this part of the task
Save all of this into a new object, e.g.
plot_data
Group data into groups with
dplyr::group_by()and ungroup data withdplyr::ungroup()Summarise each group into a single row with
dplyr::summarise()You can have multiple groups at the same time. For example:
- You can choose not to summarise, but add a new column to each group with
dplyr::mutate(). For example:
This will create a new column total_cases that contains the sum of cases for each country without reducing each group into a single row like summarise(). Values will be duplicated for each group.
tibble
- You can select rows with the top n values in a
tibbleusingdplyr::slice_max()
tibble
- You can extract values from a columns in a
tibbleusingdplyr::pull()
tibble column into a factor
- You can create and work with
factorsinside atibbleusing theforcatspackage (already intidyverse) - You can “lump”, or gather, factor levels that are not frequent with
forcats::fct_lump_n()
Task 4: Join data
Use this data
country_region.rdsto get the regions and sub-regions for all countries.Make these 2 tables:
- Write a short paragraph summarising key insights from the table, using inline R code to dynamically display case numbers. For example:
Europe (192,068) and Asia (126,420) reported the highest number of cases, while Oceania (1,879) had the lowest.
Noted that the number of cases is not hard-coded but you use inline R code to generate it within the text.
left_join()
You can join covid_cases and country_region, using the country column of covid_cases and the alpha_3 column of country_region.
You can use group_by() and summarise() to compute the total cases by sum(cases).
In the paragraph, use inline R code like {r} scales::comma(data$total_cases[tmp1$region == "Europe"]) to reference computed values. Function scales::comma() will make the numbers appear with commas for thousands (e.g., 100,000 instead of 100000).
Task 5: Data visualization
- Using
ggplot2, try your best to generate the following figure:
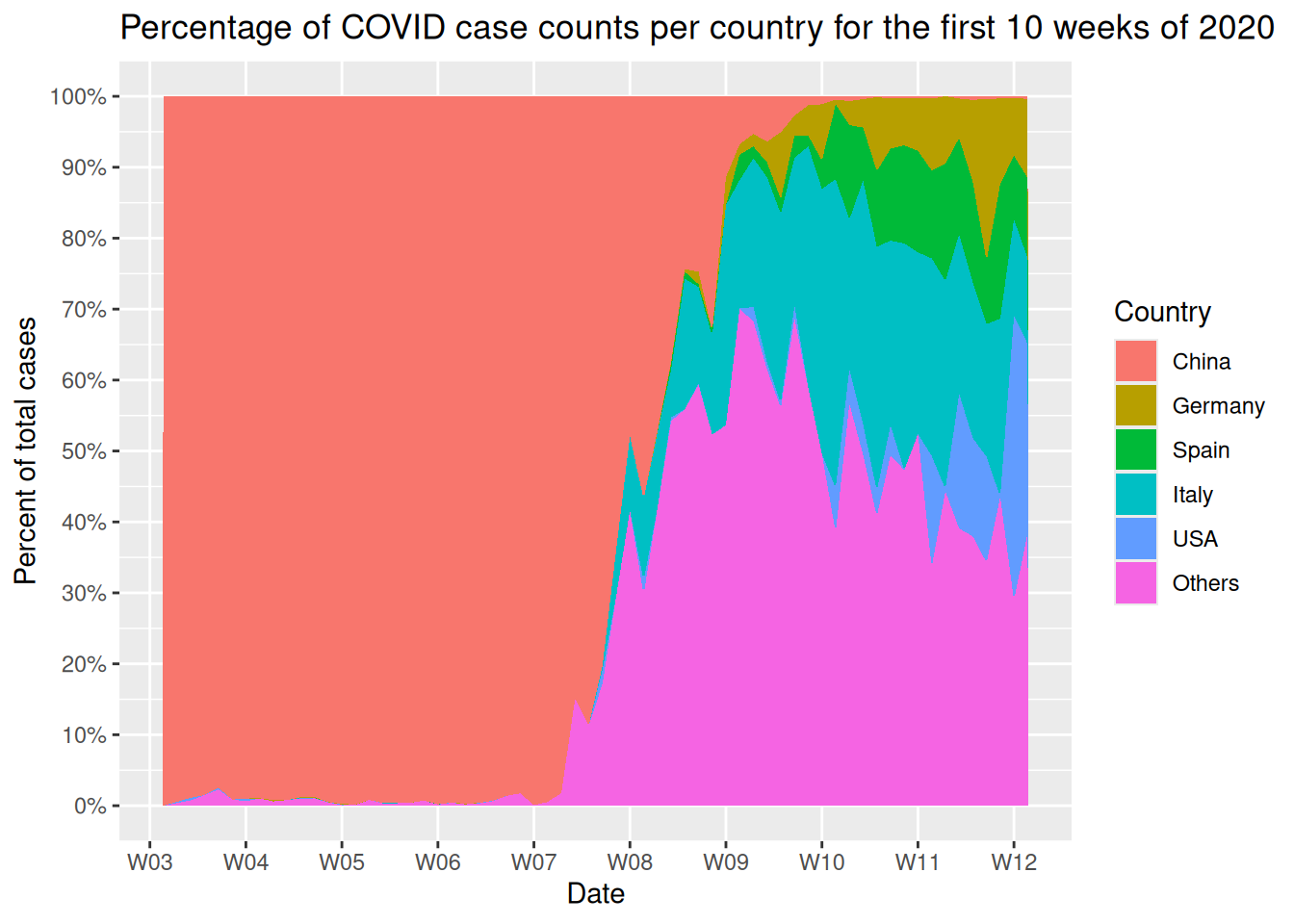
ggplot2reference page is very useful- Here are all the functions used to generate the plot above, in sequence:
ggplot()geom_area()scale_y_continuous()scale_x_date()scale_fill_discrete()ggtitle()- Every function is connected with the
+operator