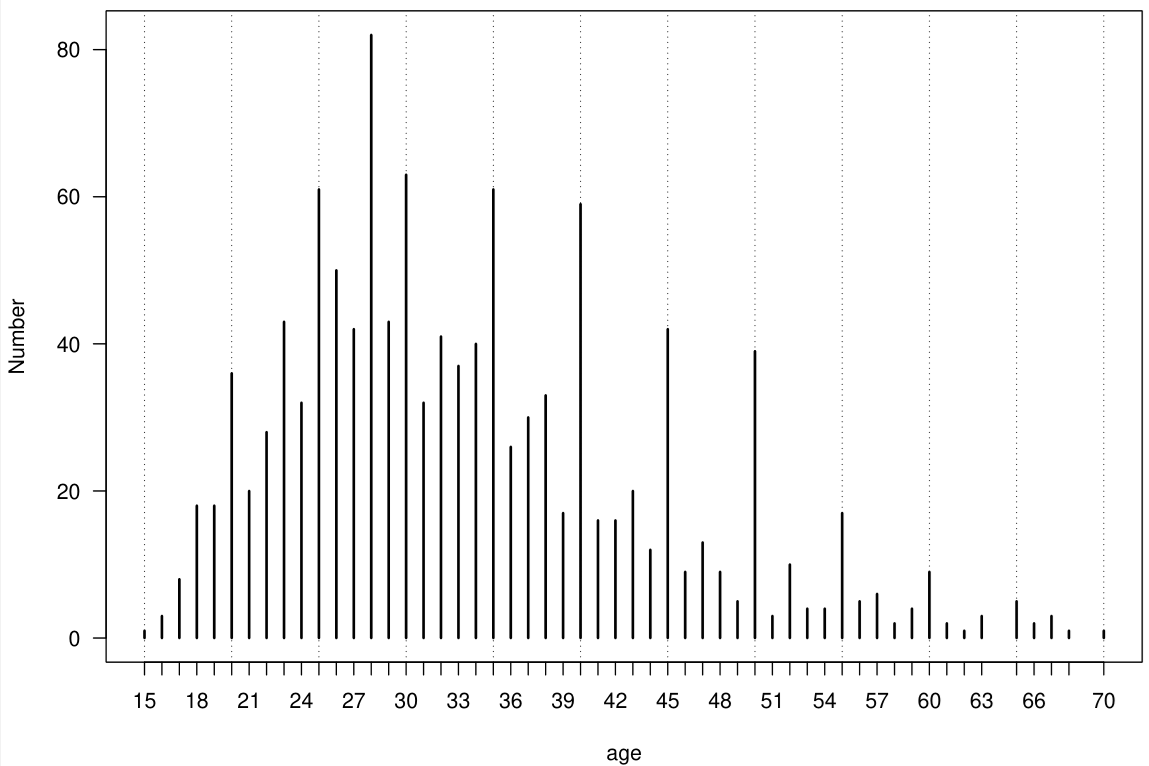
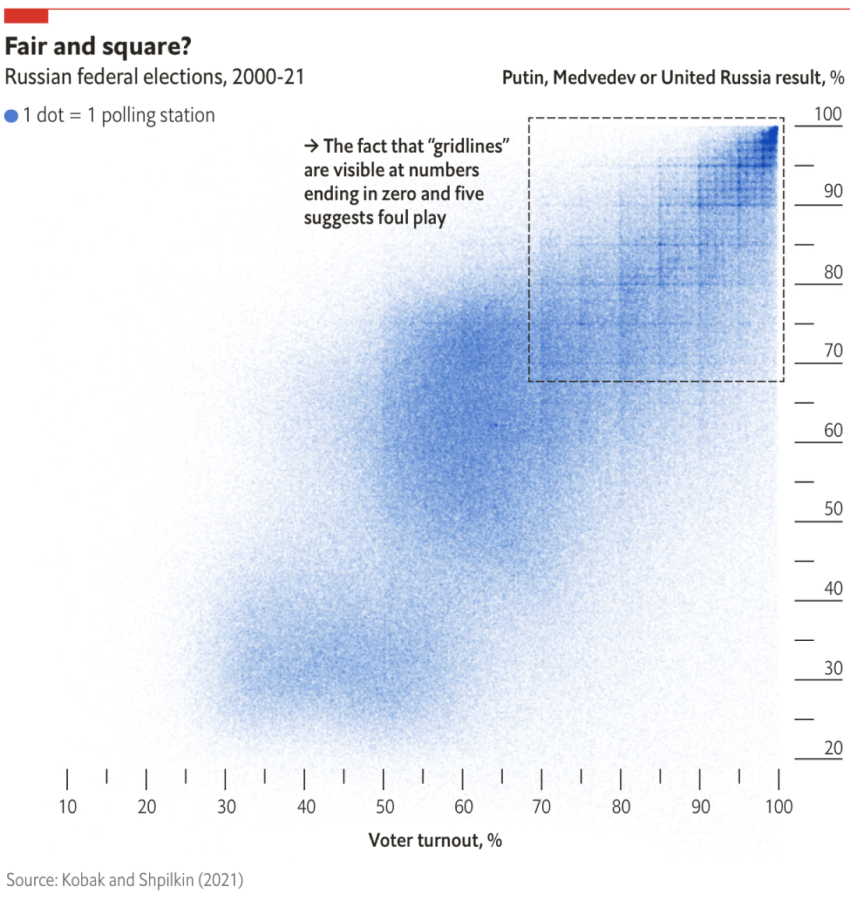
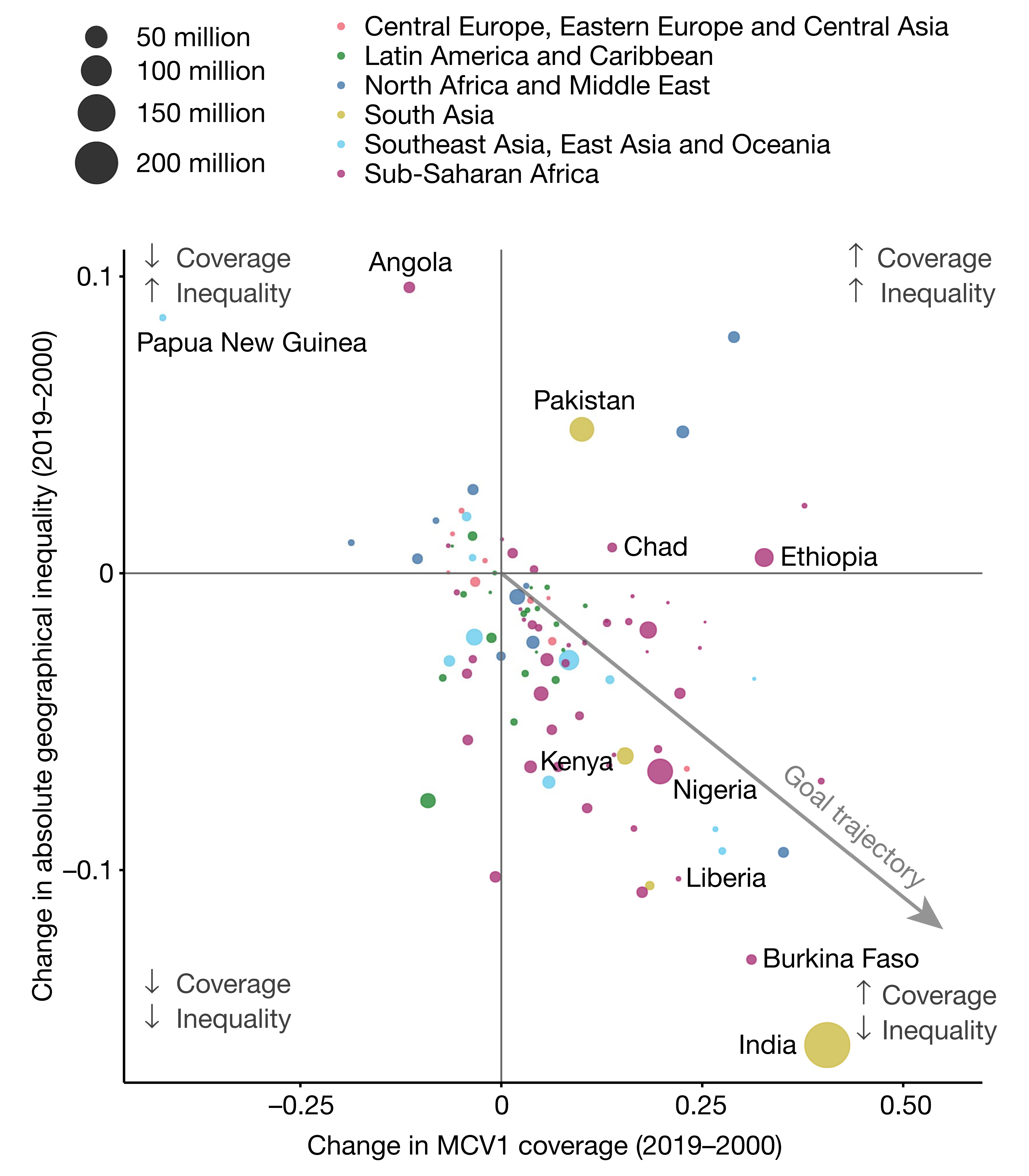
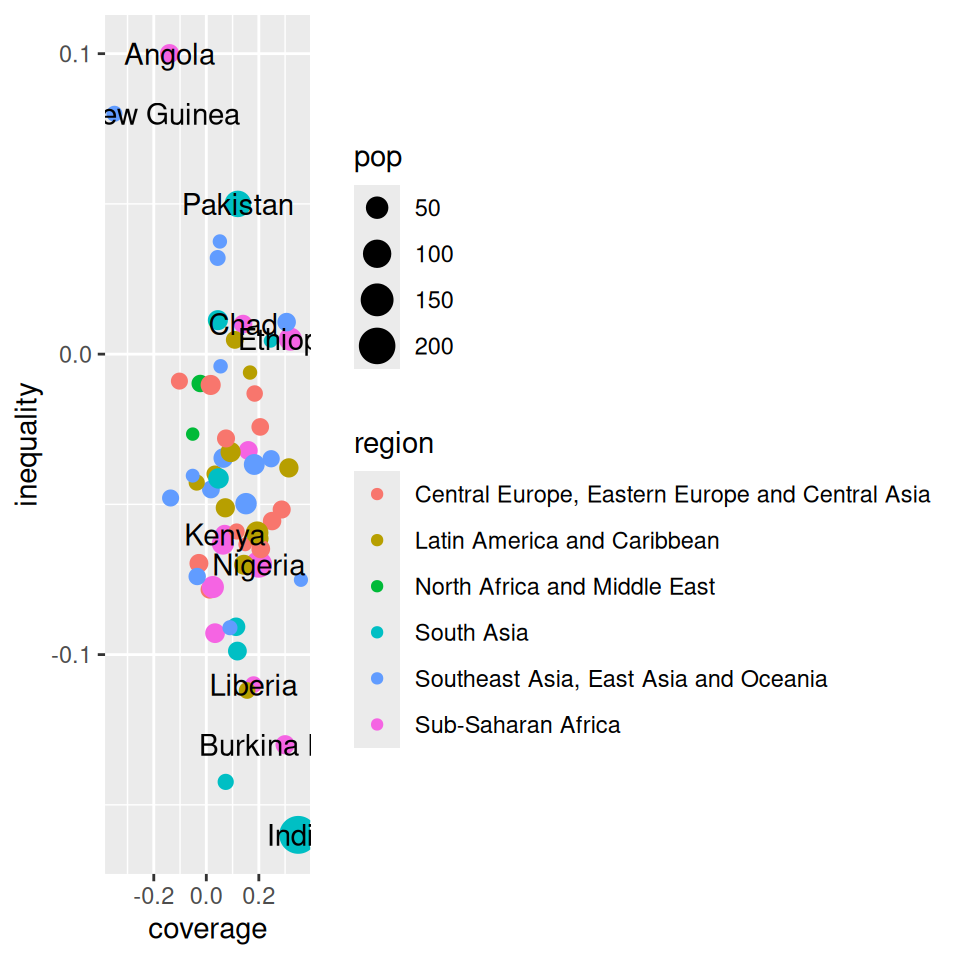
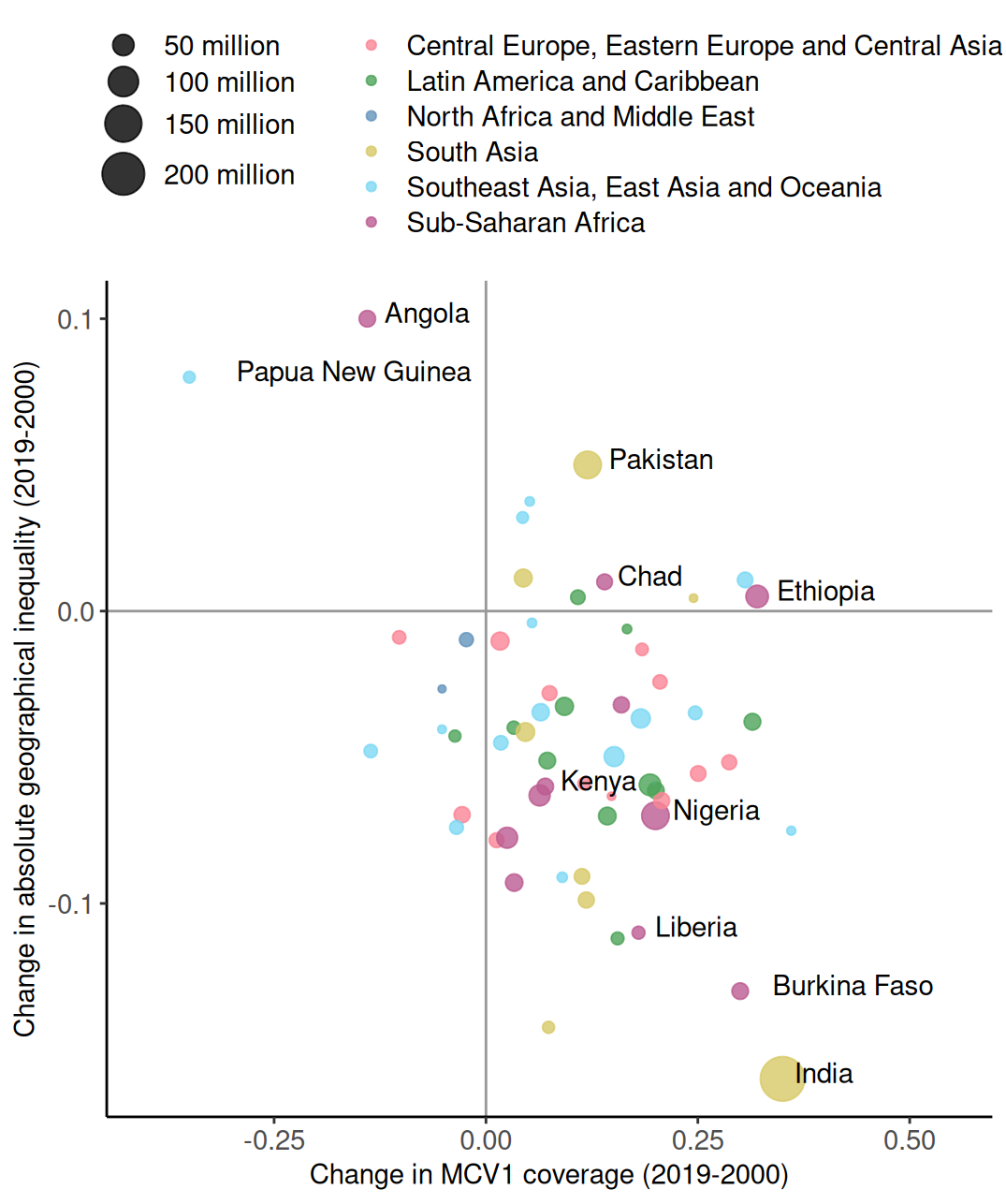
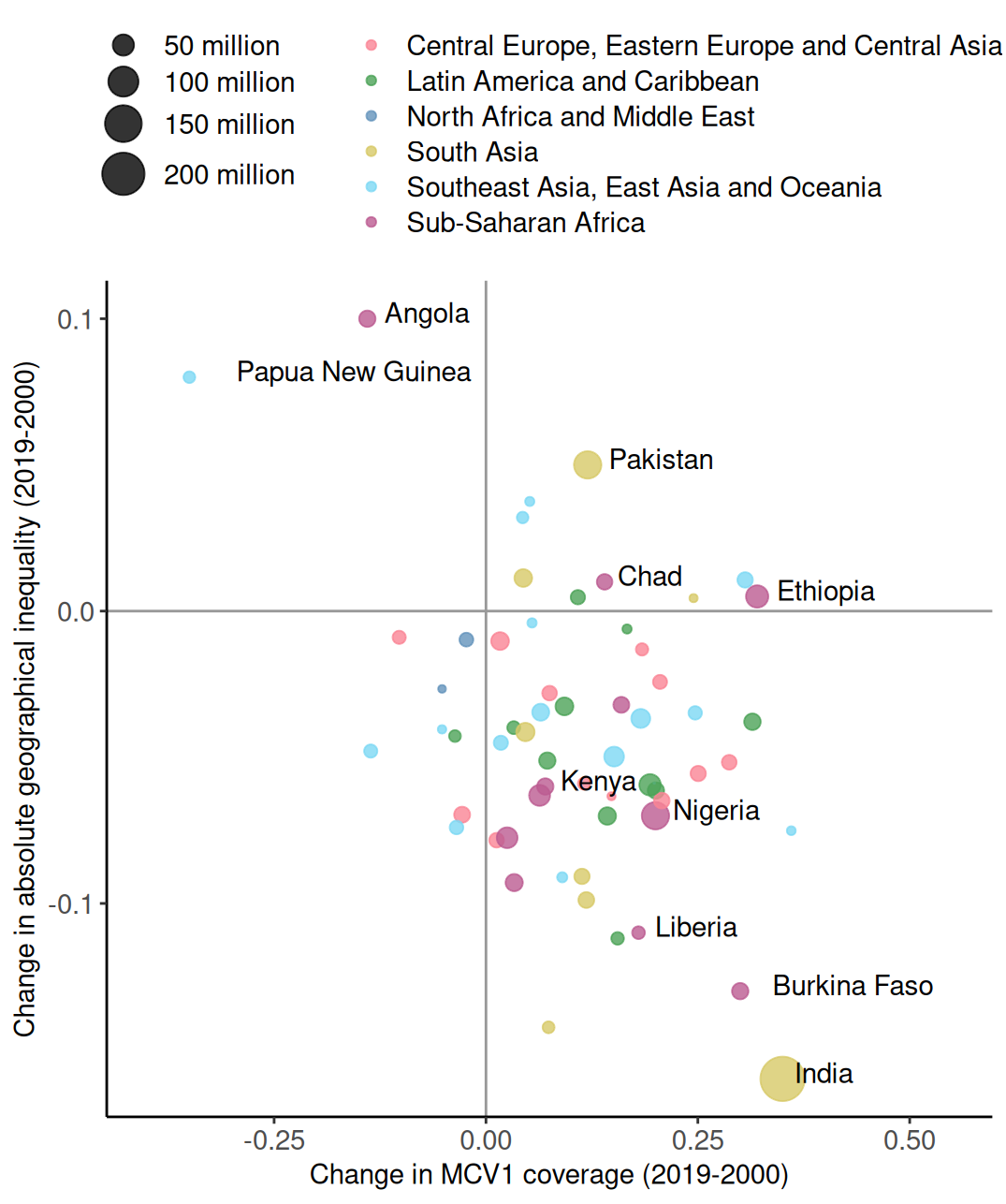
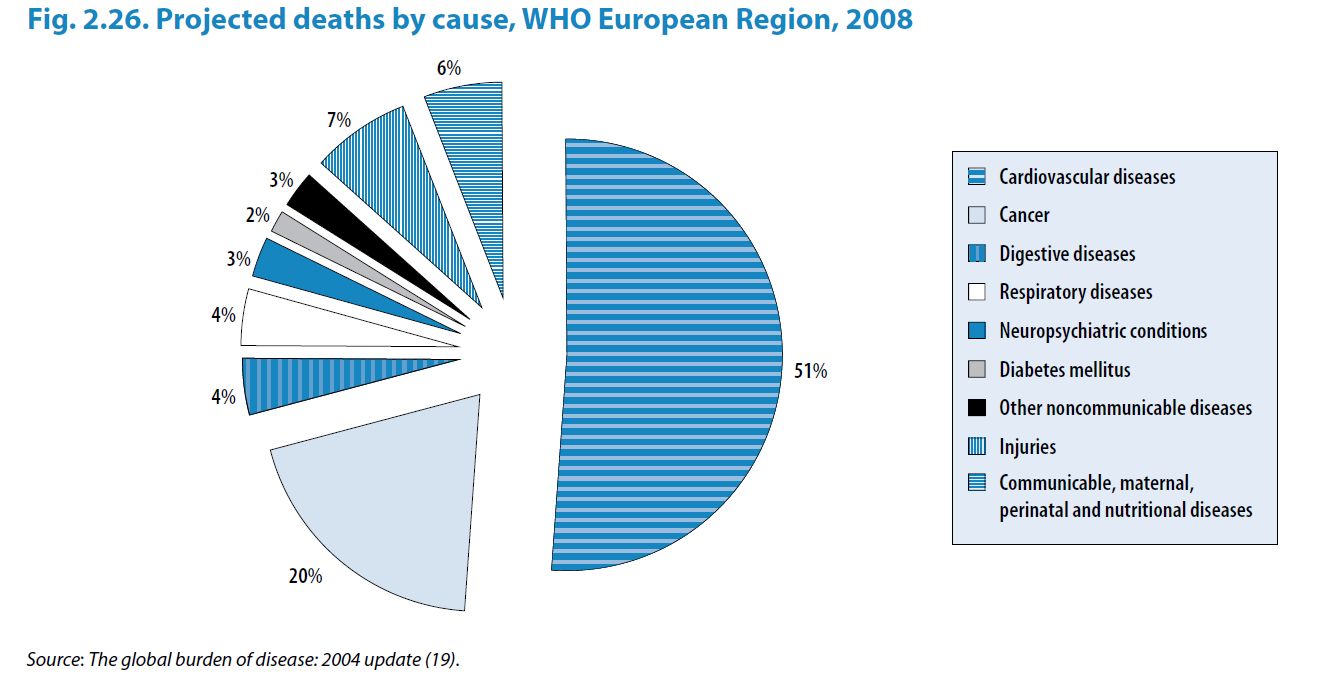
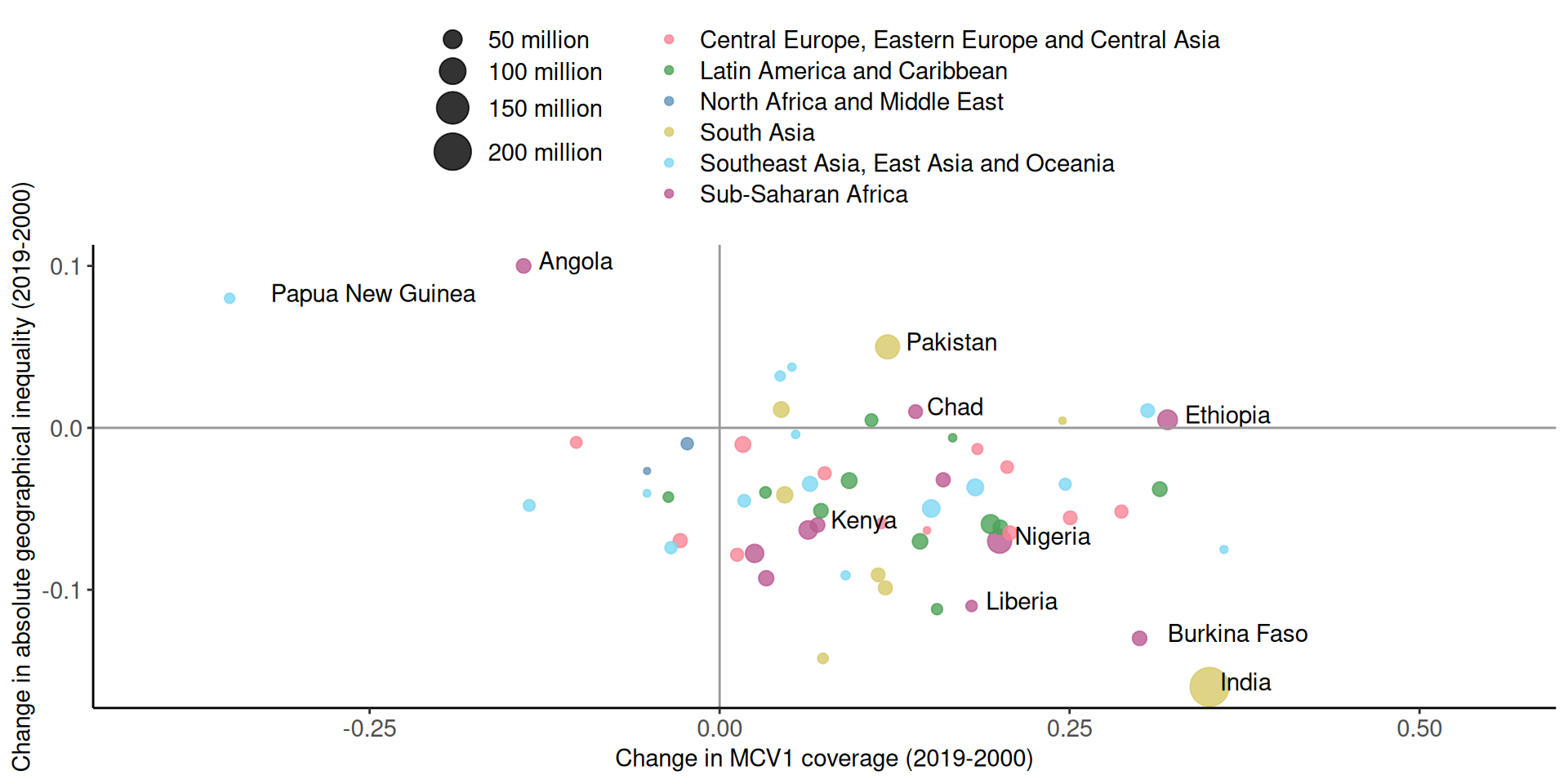
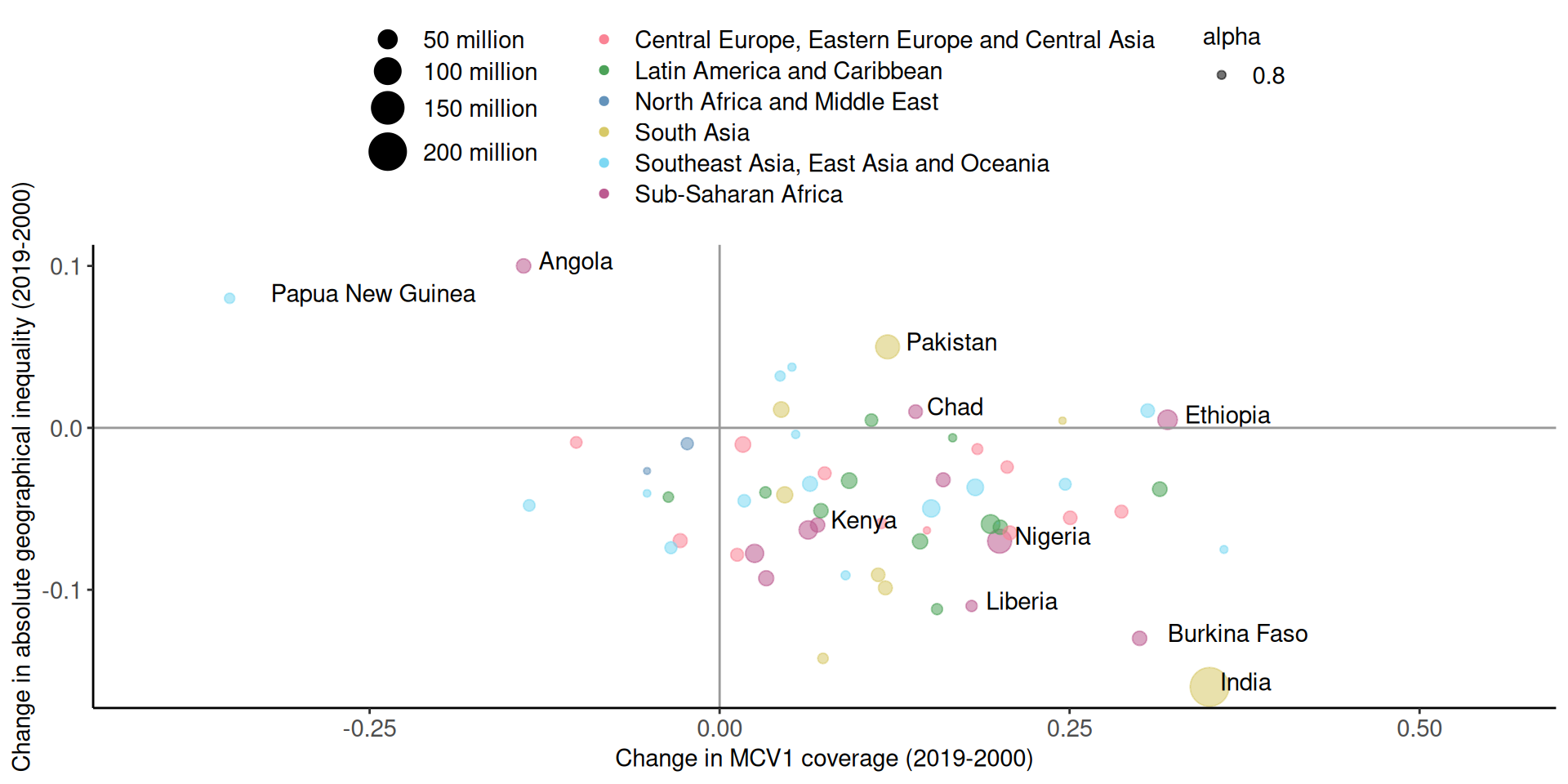
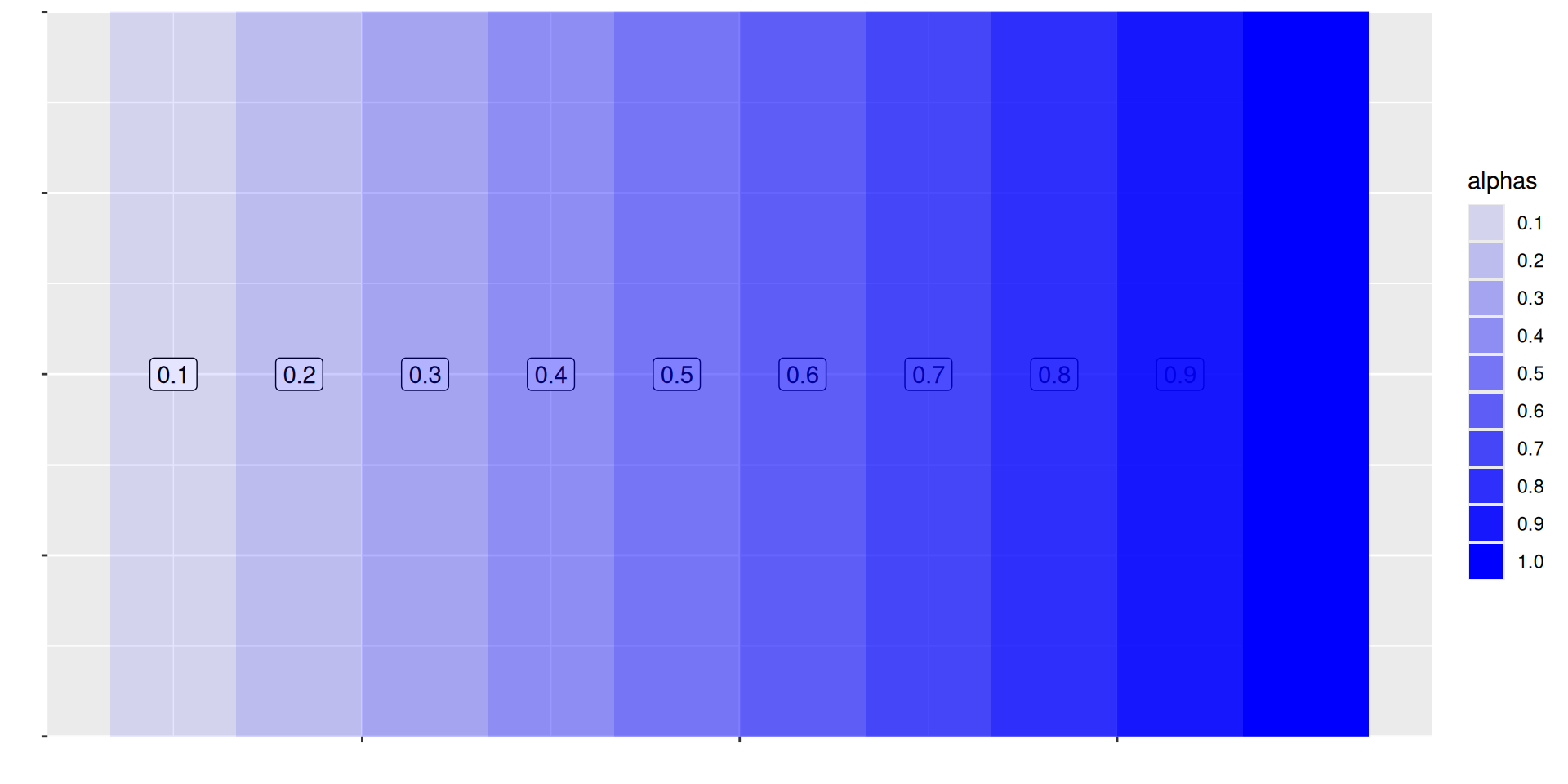
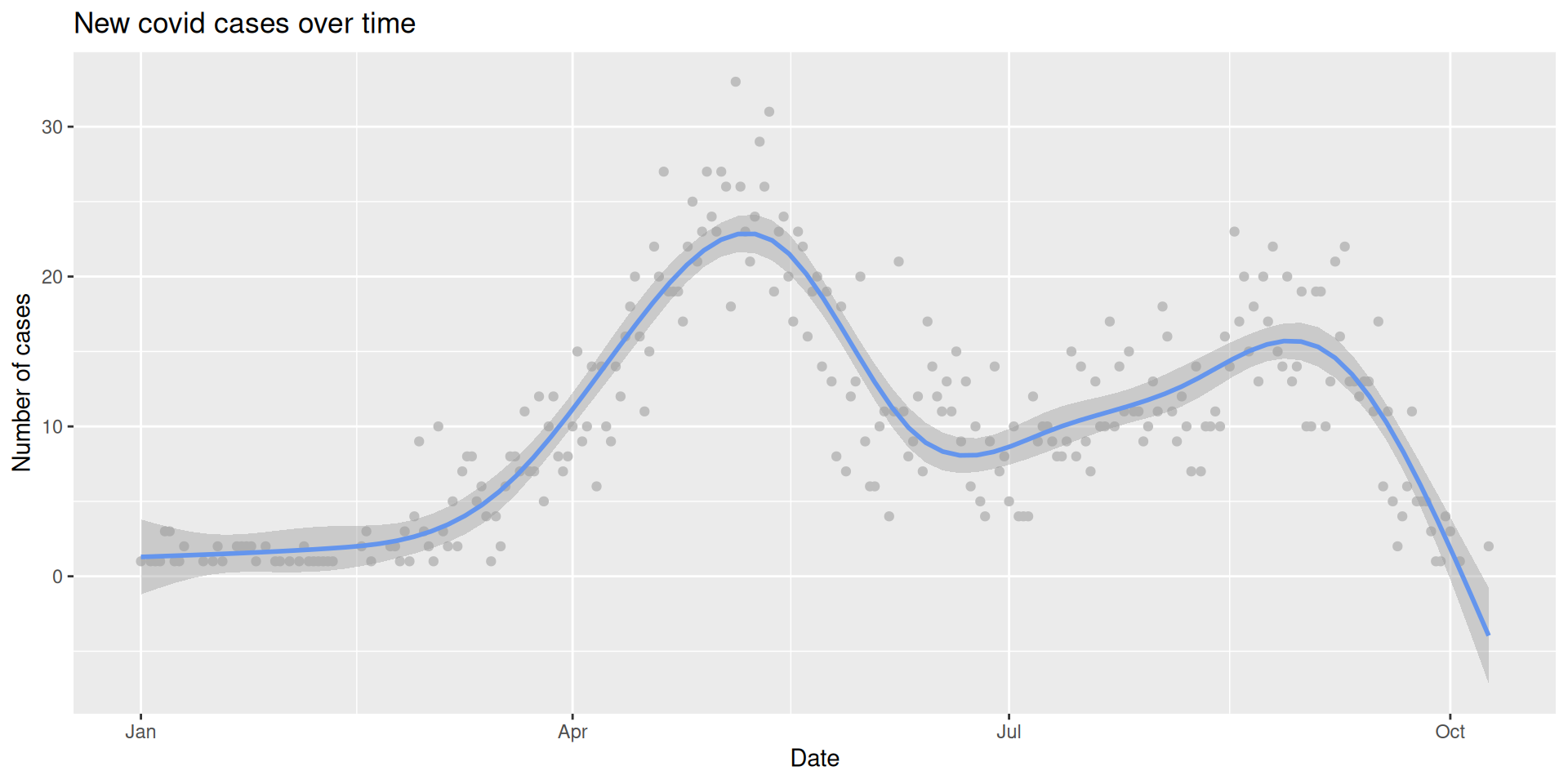
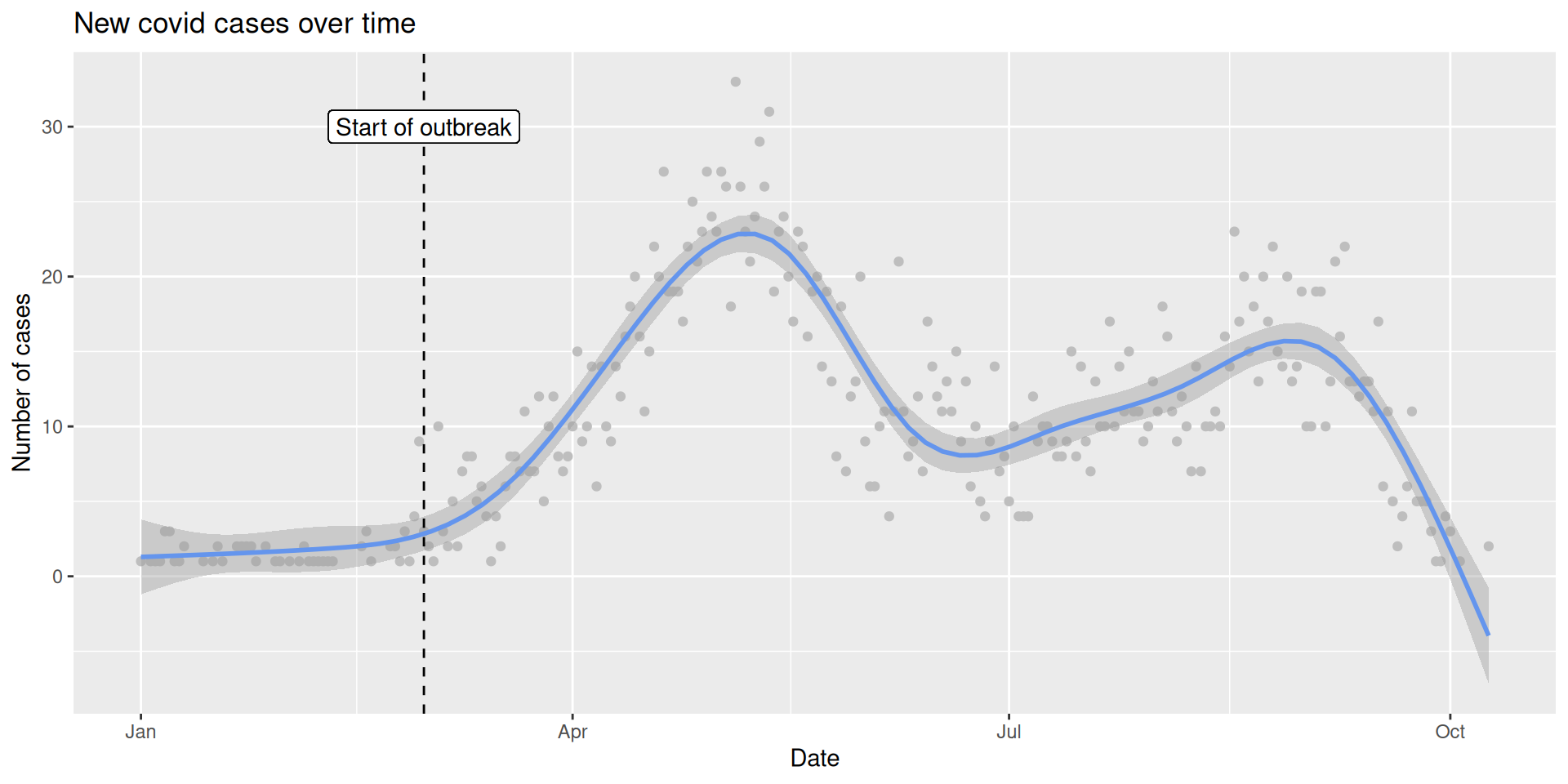
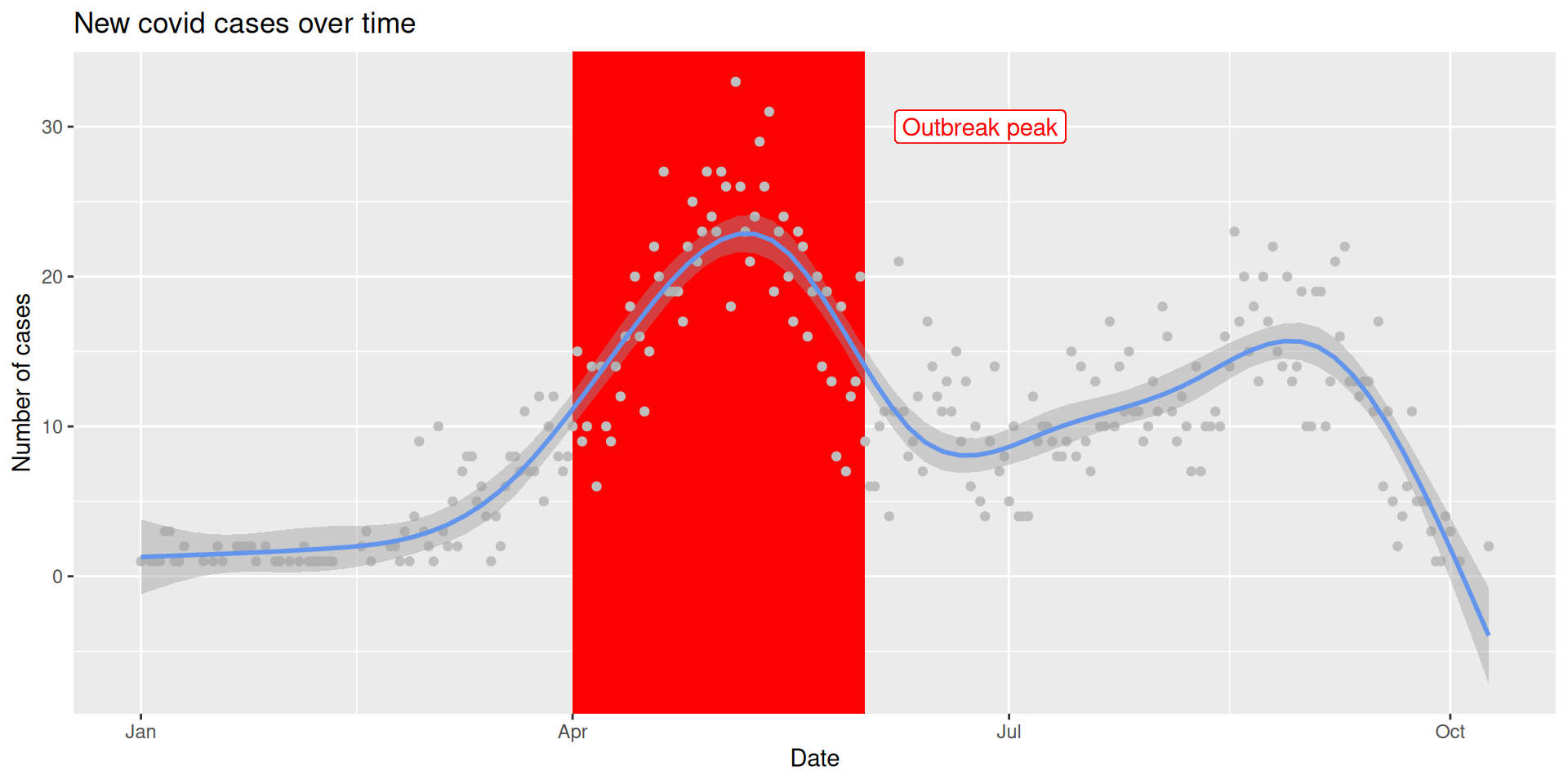
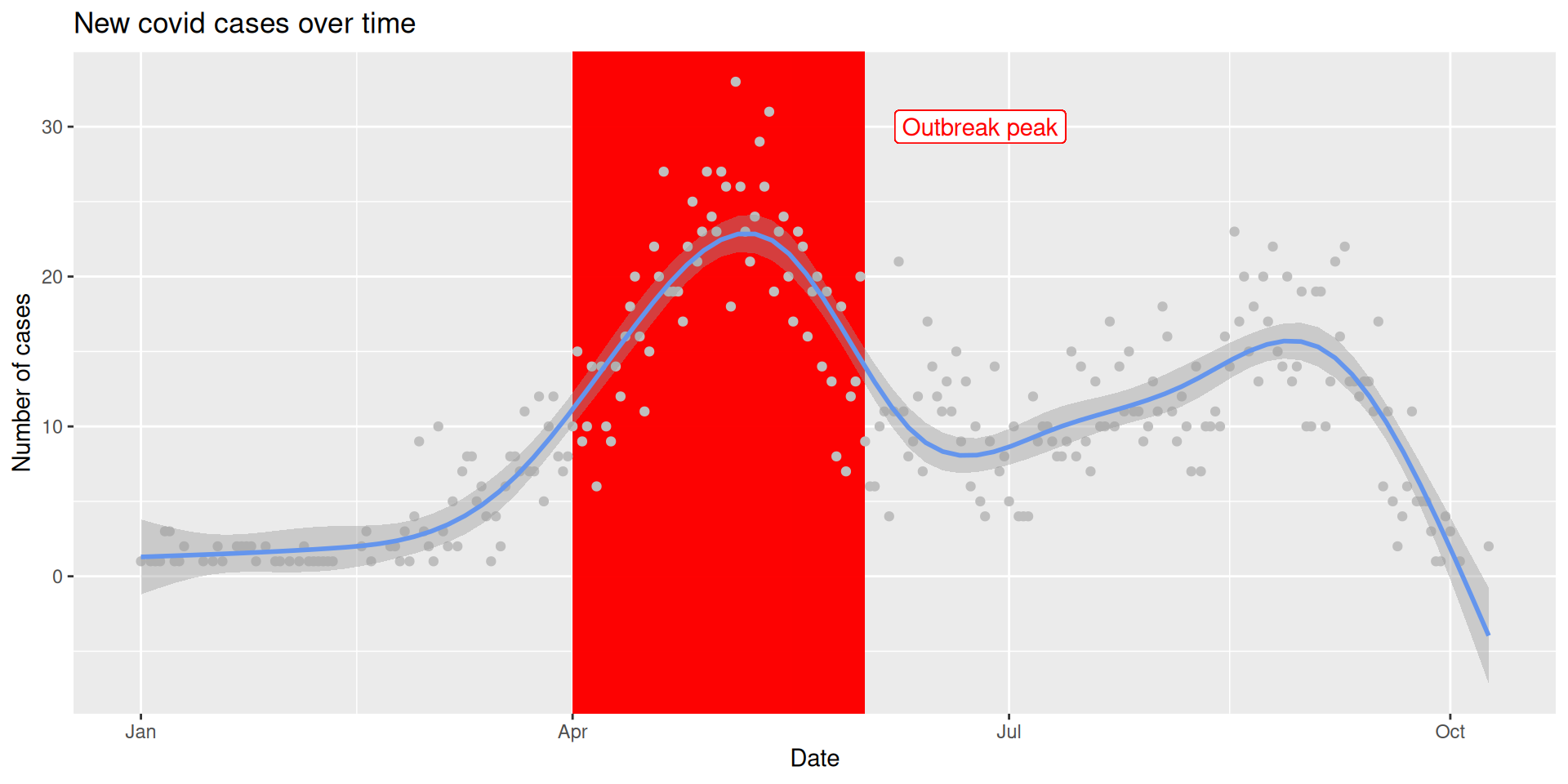
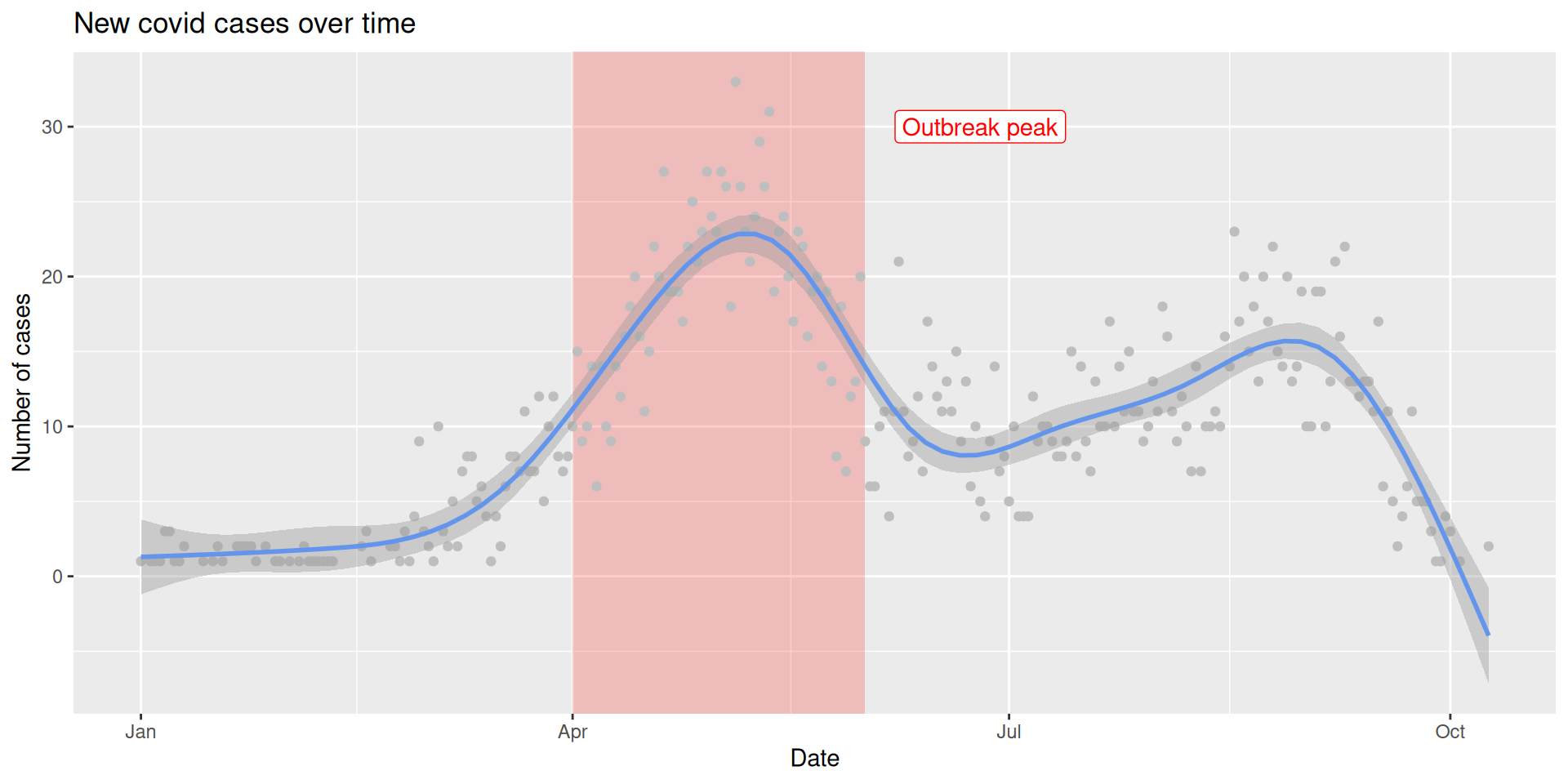
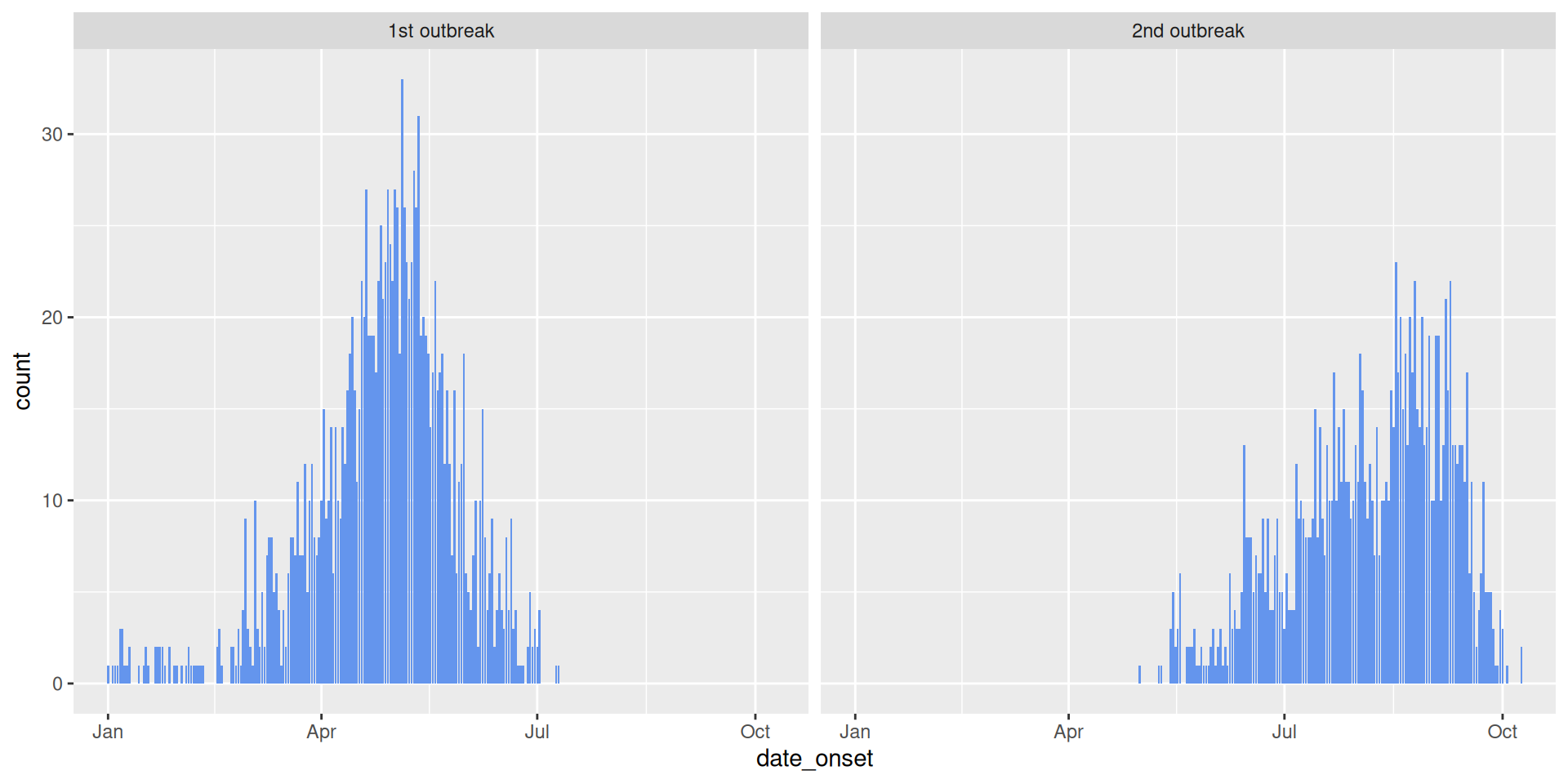
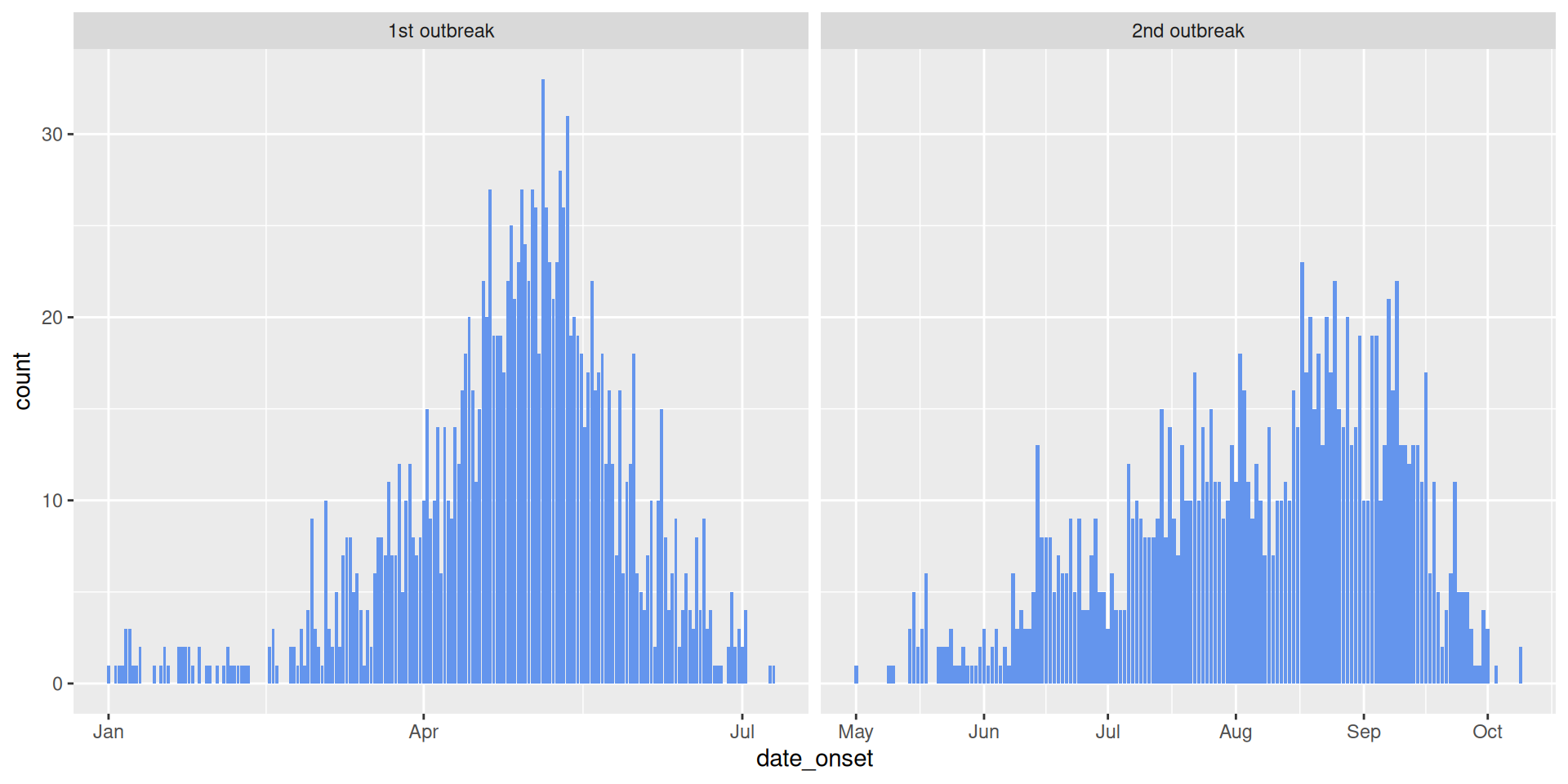
# A tibble: 6 × 5
country coverage inequality pop region
<chr> <dbl> <dbl> <dbl> <chr>
1 Angola -0.14 0.1 30.2 Sub-Saharan Africa
2 Papua New Guinea -0.35 0.08 16.5 Southeast Asia, East Asia and Ocea…
3 Pakistan 0.12 0.05 82.5 South Asia
4 Chad 0.14 0.01 27.5 Sub-Saharan Africa
5 Ethiopia 0.32 0.005 55 Sub-Saharan Africa
6 Kenya 0.07 -0.06 30.2 Sub-Saharan Africa Main components of a graph (R)
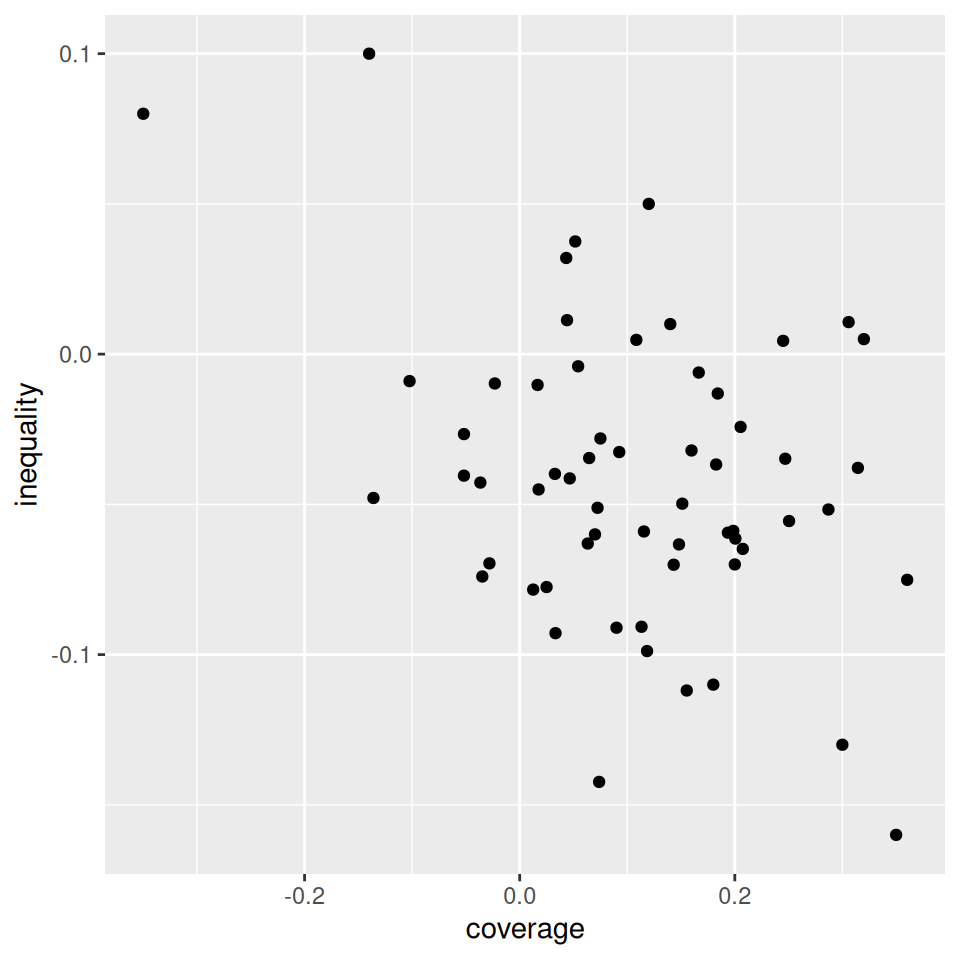
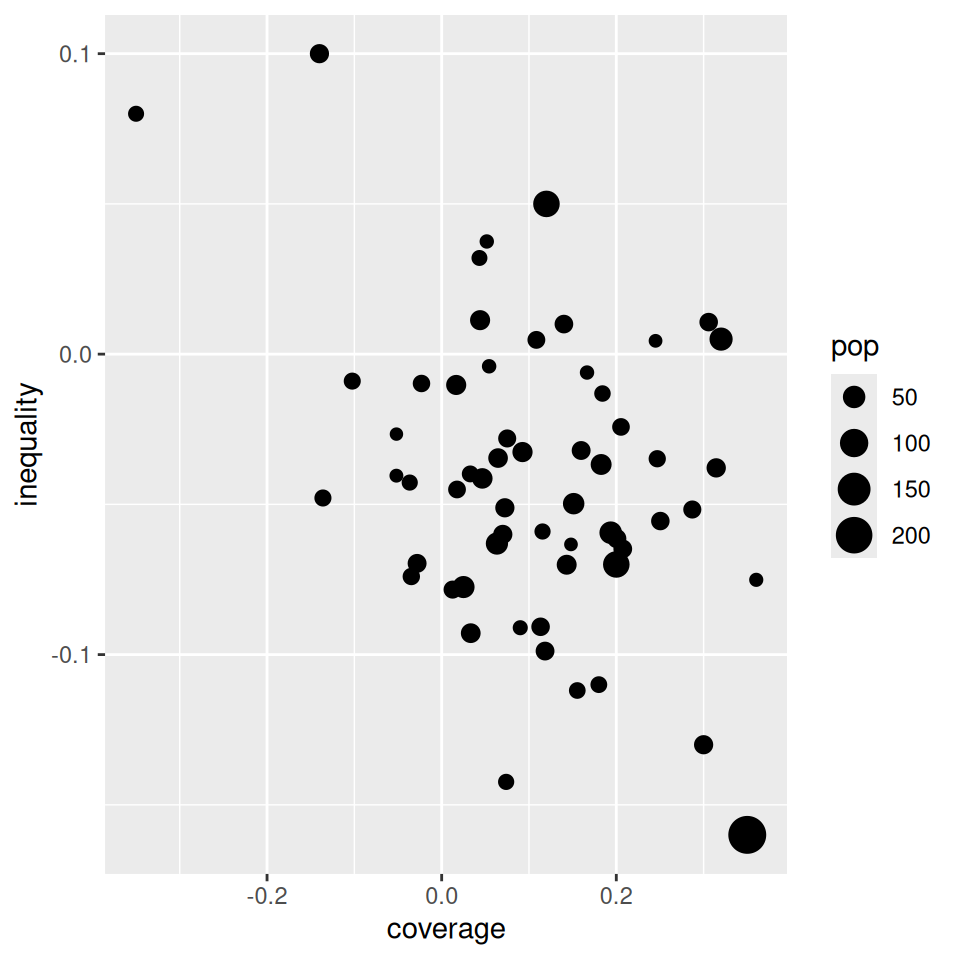
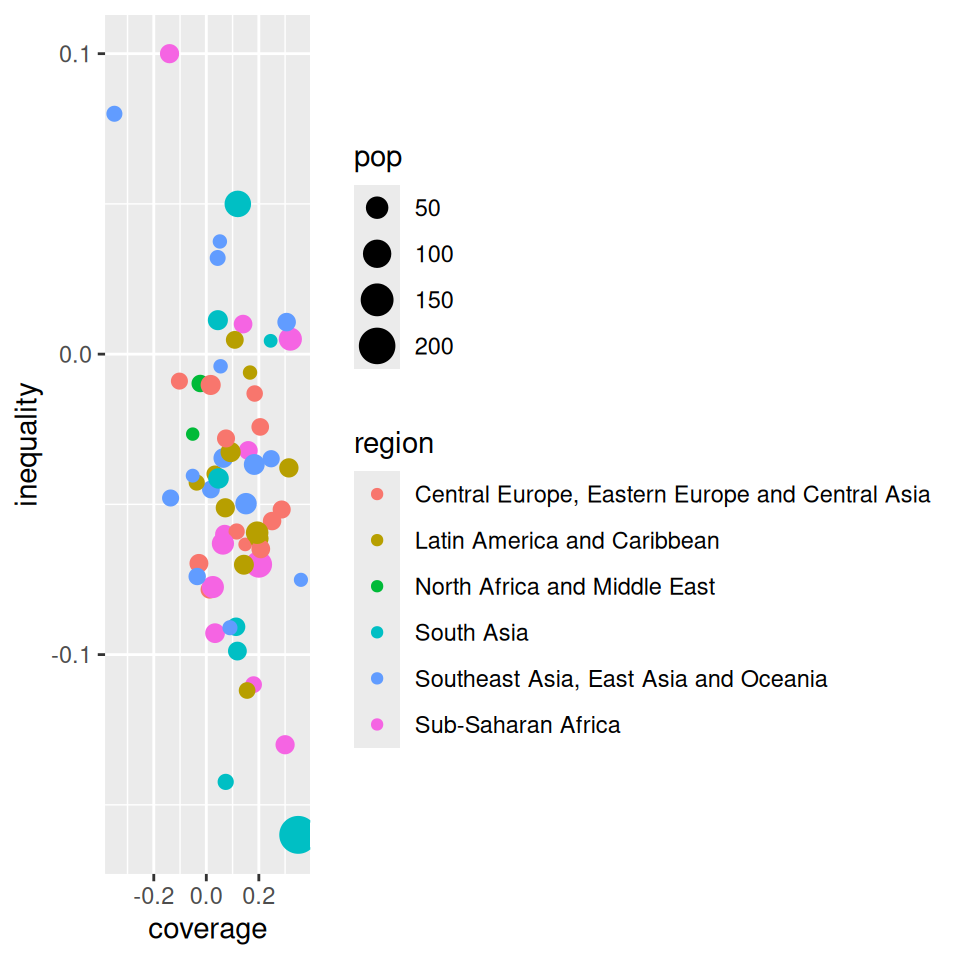
Mapping from information (data) to aesthetic attributes of geometric objects
Geometic objects: what you see on the plot (points, lines, bars)
Aesthetic attributes: “role” of variables in plot: location (often along x-axis and y-axis), colour, size, shape