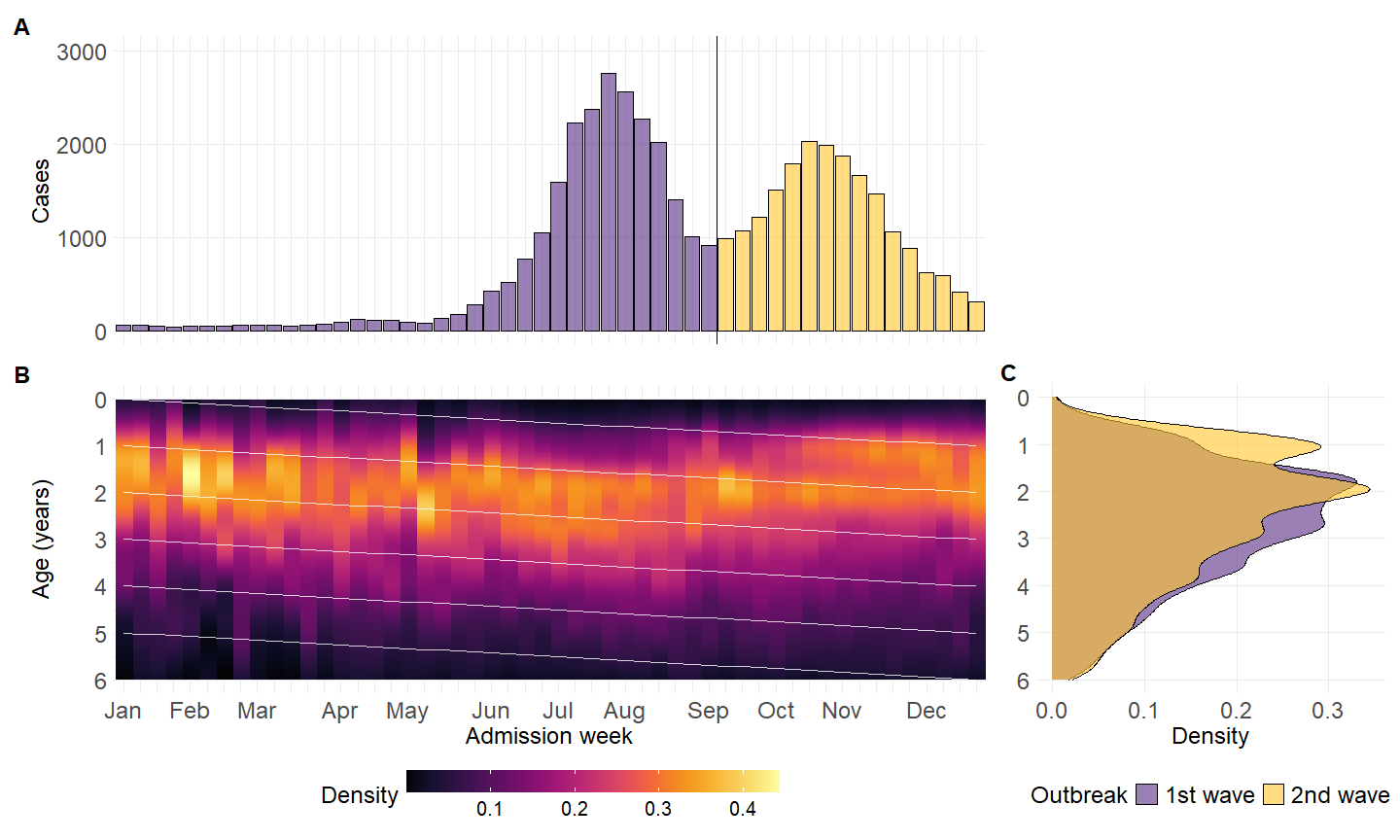
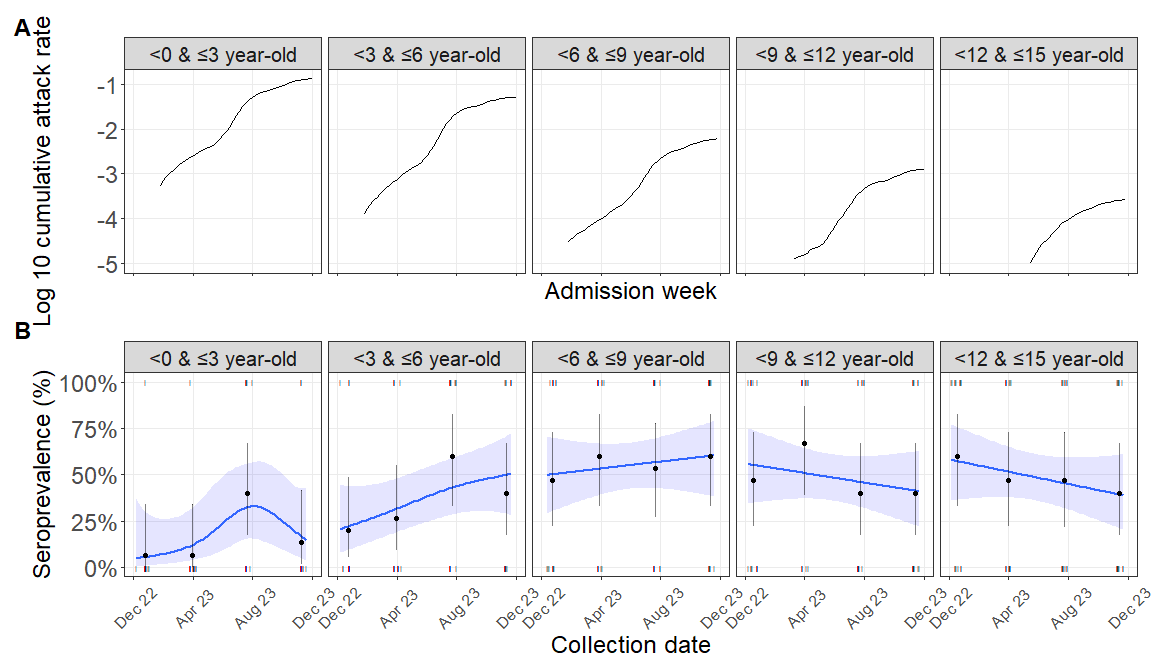
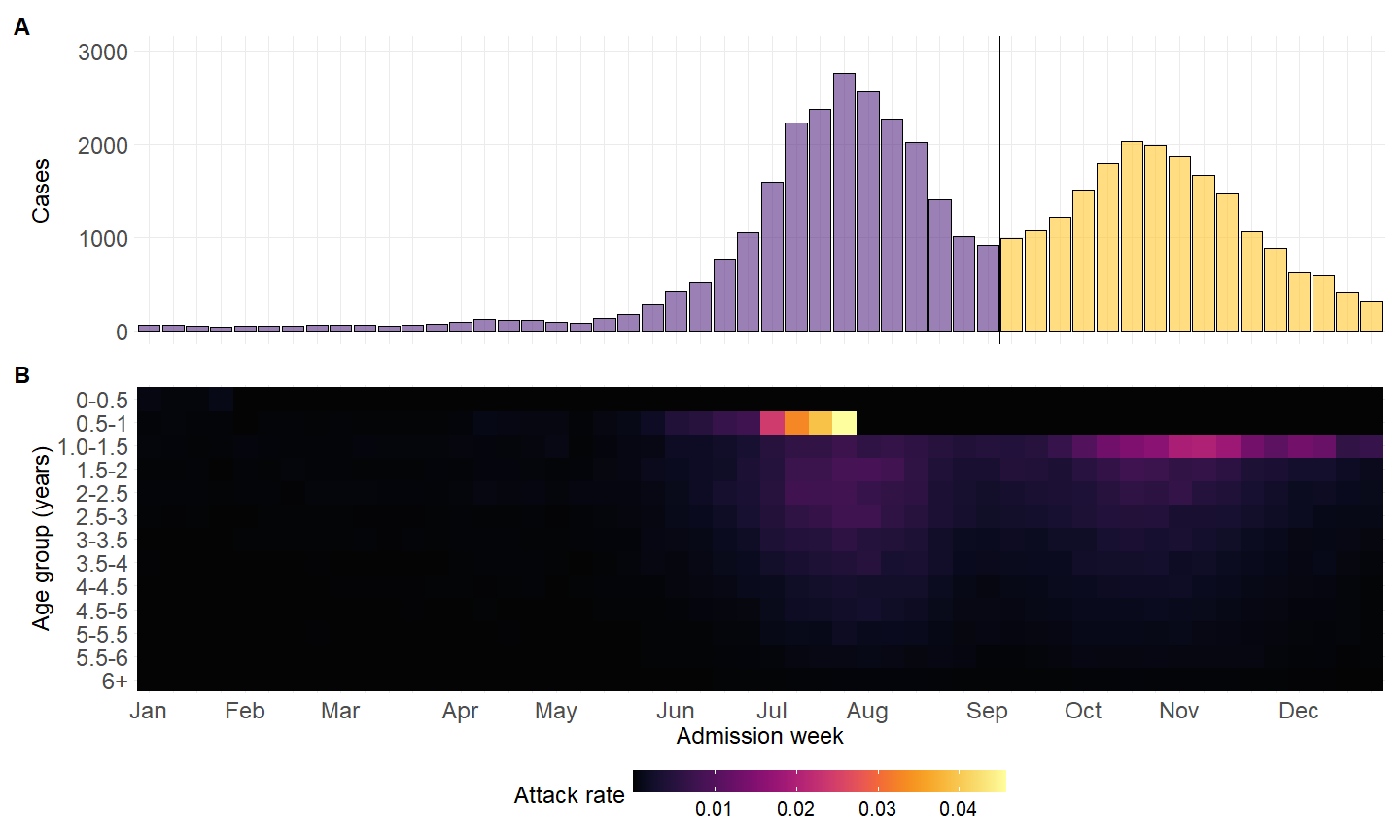
library(paletteer)
df23 <- df1 %>% filter(year(adm_week) == 2023)
co <- data.frame()
for (i in 0:6){
gen <- seq(0,1,le=52) + i
co <- rbind(co,gen)
}
df23 <- df1 %>% filter(year(adm_date) == 2023)
wwww <- slide_age(time1 = df23$adm_date,
age1 = df23$age1,
w1 = 7, s1=7)
ch <- data.frame(date = wwww$adat$date,
c0 = as.numeric(co[1,]),
c1 = as.numeric(co[2,]),
c2 = as.numeric(co[3,]),
c3 = as.numeric(co[4,]),
c4 = as.numeric(co[5,]),
c5 = as.numeric(co[6,]))
leb_month <- c("Jan",rep("",3),"Feb",rep("",3),"Mar",rep("",4),"Apr",rep("",3),
"May",rep("",4),"Jun",rep("",3),"Jul",rep("",3),"Aug",rep("",4),
"Sep",rep("",3),"Oct",rep("",3),"Nov",rep("",4),"Dec",rep("",3))
ts <- data.frame(wwww$wdat$date,wwww$wdat$age) %>%
filter(!is.na(wwww$wdat$date) & !is.na(wwww$wdat$age)) %>%
count(wwww$wdat$date) %>%
set_colnames(c("time","n")) %>%
mutate(peak = c(rep("1st",36),rep("2nd",nrow(.)-36))) %>%
ggplot(aes(x = time, y = n,fill = peak)) +
geom_col(alpha = 0.6,color="black") +
geom_vline(xintercept = 36.5) +
theme_minimal()+
scale_y_continuous(name = "Cases",
breaks = seq(0,3000,by = 1000),
limit =c(0,3000),
minor_breaks = NULL)+
scale_fill_manual(values = c("#582C83FF","#FFC72CFF"))+
labs(tag = "A")+
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "none",
plot.tag = element_text(face = "bold", size = 18),
axis.title.y = element_text(size = 18),
axis.text.y = element_text(size = 18))
hm <- ggplot(data=wwww$wdat, aes(x=date, y=age)) +
stat_density(
aes(fill = after_stat(density)),
geom = "raster",
position = "identity"
)+
scale_fill_paletteer_c("grDevices::Inferno")+
# scale_fill_gradient(low="#040404FF", high= "#FFFE9EFF")+
# scale_fill_distiller(palette = "Blues")+
theme_minimal()+
scale_y_reverse(name = "Age (years)",lim= rev(c(0,6)),breaks = seq(0,6))+
scale_x_discrete(name = "Admission week",labels = leb_month)+
labs(tag = "B",fill = "Density")+
# theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1,size = 8))+
geom_line(data = ch,aes(x = date,y = c0),col = "white",
group = 1,lwd = 0.25,alpha = 0.8)+
geom_line(data = ch,aes(x = date,y = c1),col = "white",
group = 1,lwd = 0.25,alpha = 0.8)+
geom_line(data = ch,aes(x = date,y = c2),col = "white",
group = 1,lwd = 0.25,alpha = 0.8)+
geom_line(data = ch,aes(x = date,y = c3),col = "white",
group = 1,lwd = 0.25,alpha = 0.8)+
geom_line(data = ch,aes(x = date,y = c4),col = "white",
group = 1,lwd = 0.25,alpha = 0.8)+
geom_line(data = ch,aes(x = date,y = c5),col = "white",
group = 1,lwd = 0.25,alpha = 0.8)+
theme(axis.title.y = element_text(size = 18),
axis.ticks.x = element_blank(),
legend.position = "bottom",
plot.tag = element_text(face = "bold", size = 18),
axis.title.x = element_text(size = 18),
axis.text.x = element_text(size = 18),
axis.text.y = element_text(size = 18),
legend.text = element_text(size = 15),
legend.title = element_text(size = 18))+
guides(fill=guide_colourbar(barwidth=20,label.position="bottom"))
fi_peak <- df1 %>% filter(year(adm_date) == "2023") %>%
filter((adm_date <= as.Date("2023-09-03")&
!is.na(adm_date) & !is.na(age1)))
se_peak <- df1 %>% filter(year(adm_date) == "2023") %>%
filter((adm_date > as.Date("2023-09-03")) &
!is.na(adm_date) & !is.na(age1))
data <- data.frame(
peak = c( rep("1st wave",nrow(data.frame(se_peak$age1))),
rep("2nd wave",nrow(data.frame(fi_peak$age1)))),
age = c( fi_peak$age1, se_peak$age1 )
)
## density plot
ad <- ggplot(data=data, aes(x=age, group=peak, fill=peak)) +
geom_density(alpha = 0.6) +
scale_fill_manual(values = c("#582C83FF","#FFC72CFF")) +
scale_x_reverse(limit = c(6,0),breaks = seq(0,6,by=1),
minor_breaks = NULL)+
scale_y_continuous(minor_breaks = NULL)+
coord_flip()+
theme_minimal()+
labs(x = "Age", y ="Density",tag = "C",fill = "Outbreak")+
theme(axis.title.y = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "bottom",
plot.tag = element_text(face = "bold", size = 18),
axis.title.x = element_text(size = 18),
axis.text.x = element_text(size = 18),
axis.text.y = element_text(size = 18),
legend.text = element_text(size = 18),
legend.title = element_text(size = 18))