library(paletteer)
df23 <- df1 %>% filter(year(adm_week) == 2023)
co <- data.frame()
for (i in 0:6){
gen <- seq(0,1,le=52) + i
co <- rbind(co,gen)
}
df23 <- df1 %>% filter(year(adm_date) == 2023)
wwww <- slide_age(time1 = df23$adm_date,
age1 = df23$age1,
w1 = 7, s1=7)
library(zoo)
ch <- data.frame(date = wwww$adat$date,
c0 = as.numeric(co[1,]),
c1 = as.numeric(co[2,]),
c2 = as.numeric(co[3,]),
c3 = as.numeric(co[4,]),
c4 = as.numeric(co[5,]),
c5 = as.numeric(co[6,]))
ch <- ch %>% mutate(
trend = c(rep("#80FFFFFF",26-3),
rep("#FFFFFFFF",26+3))
)
ch$group <- consecutive_id(ch$trend)
ch <- head(do.call(rbind, by(ch, ch$group, rbind, NA)), -1)
ch[, c("trend", "group")] <- lapply(ch[, c("trend", "group")], na.locf)
ch[] <- lapply(ch, na.locf, fromLast = T)
ch$trend <- factor(ch$trend,
levels = c("#80FFFFFF","#FFFFFFFF"))
leb_month <- c("Jan",rep("",3),"Feb",rep("",3),"Mar",rep("",4),"Apr",rep("",3),
"May",rep("",4),"Jun",rep("",3),"Jul",rep("",3),"Aug",rep("",4),
"Sep",rep("",3),"Oct",rep("",3),"Nov",rep("",4),"Dec",rep("",3))
ts <- data.frame(wwww$wdat$date,wwww$wdat$age) %>%
filter(!is.na(wwww$wdat$date) & !is.na(wwww$wdat$age)) %>%
count(wwww$wdat$date) %>%
set_colnames(c("time","n")) %>%
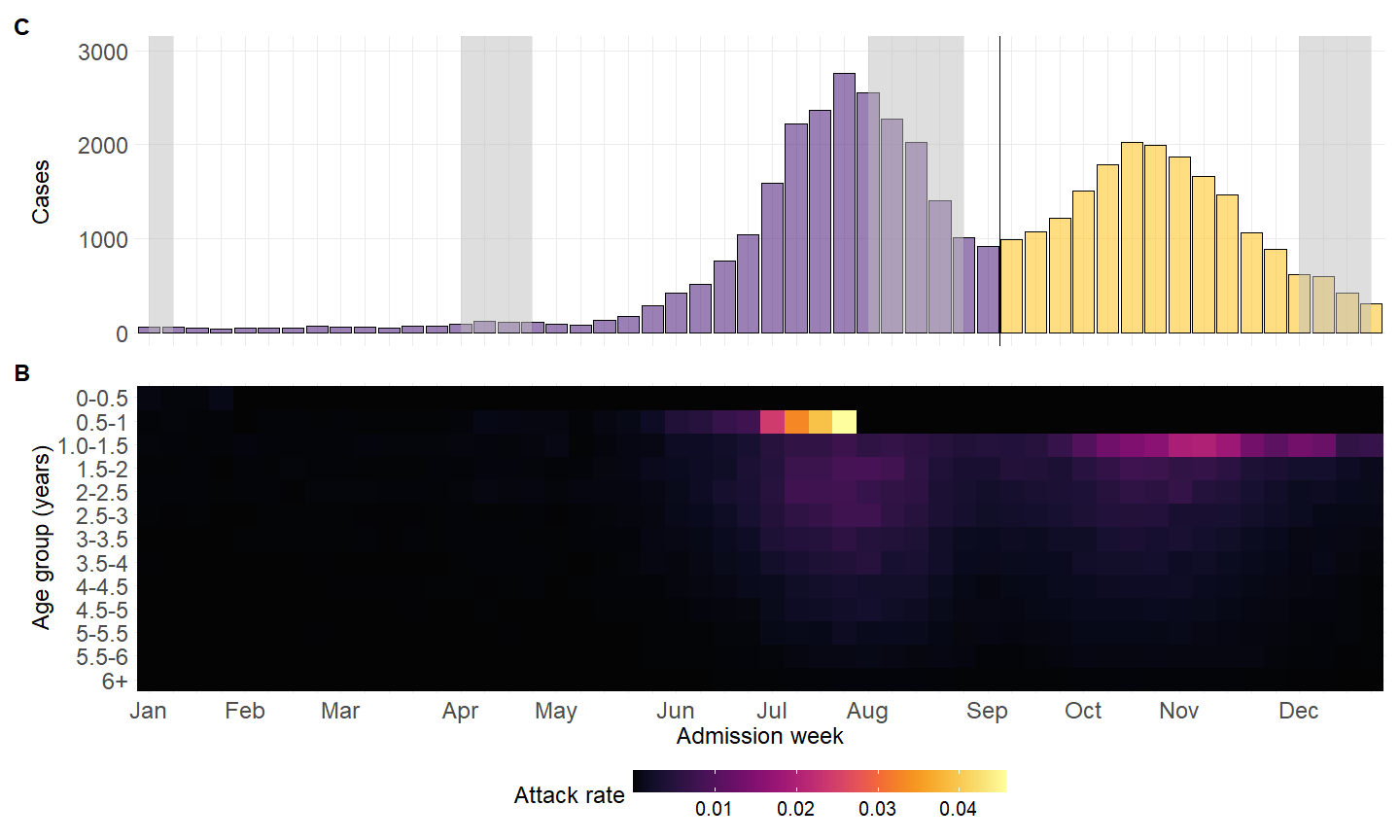
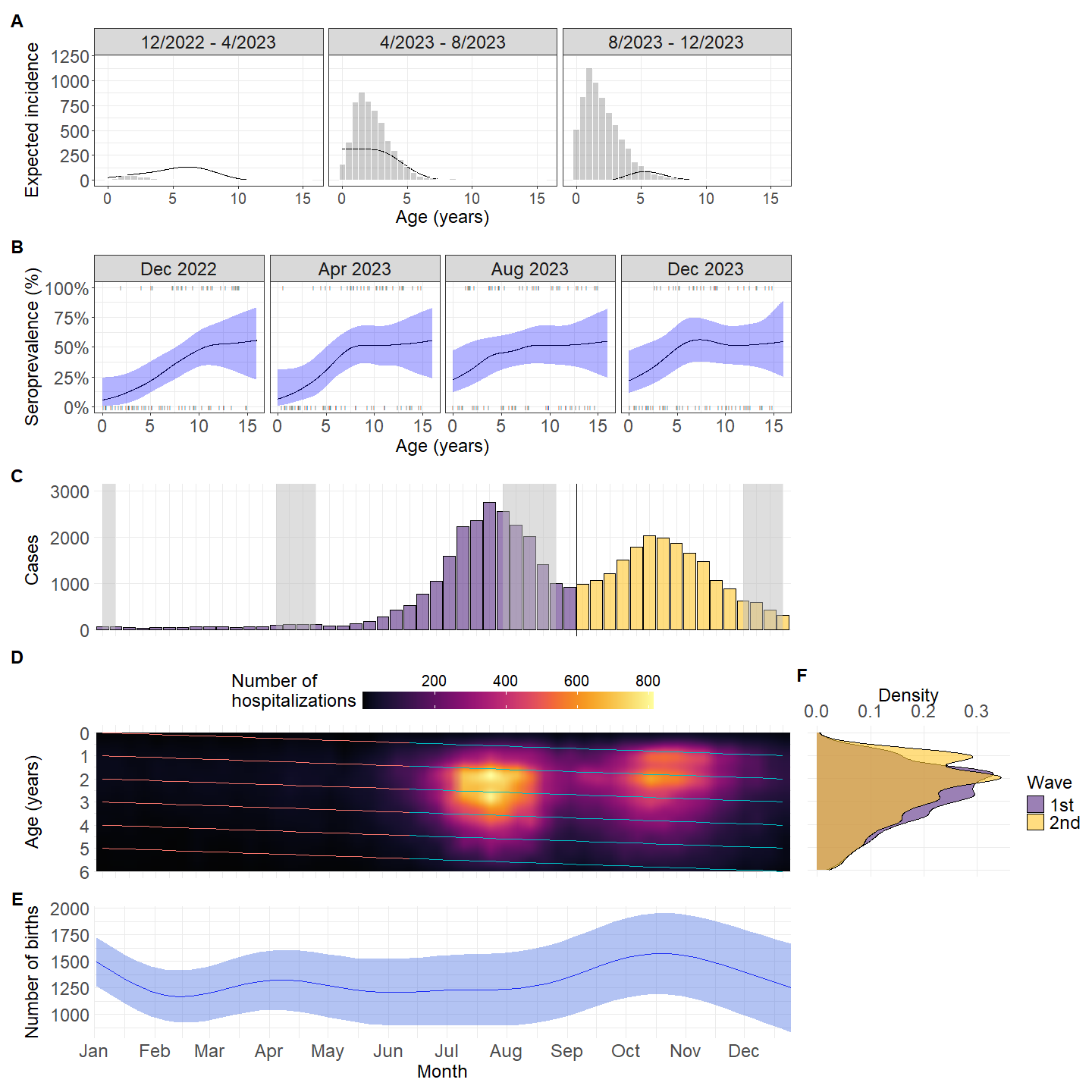
mutate(peak = c(rep("1st",36),rep("2nd",nrow(.)-36))) %>%
ggplot(aes(x = time, y = n,fill = peak)) +
geom_col(alpha = 0.6,color="black") +
geom_vline(xintercept = 36.5) +
theme_minimal()+
scale_y_continuous(name = "Cases",
breaks = seq(0,3000,by = 1000),
limit =c(0,3000),
minor_breaks = NULL)+
scale_fill_manual(values = c("#582C83FF","#FFC72CFF"))+
labs(tag = "C")+
annotate("rect", fill = "grey",
xmin = c("2023-01-04","2023-04-05","2023-08-02","2023-12-06"),
xmax = c("2023-01-11","2023-04-26","2023-08-30","2023-12-27"),
ymin = 0, ymax = Inf, alpha = .5)+
theme(axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "none",
plot.tag = element_text(face = "bold", size = 18),
axis.title.y = element_text(size = 18),
axis.text.y = element_text(size = 18))
hm <- ggplot(data=wwww$wdat, aes(x=date, y=age)) +
stat_density(
aes(fill = after_stat(density)),
geom = "raster",
position = "identity",
interpolate = TRUE
)+
scale_fill_paletteer_c("grDevices::Inferno")+
# scale_fill_gradient(low="#040404FF", high= "#FFFE9EFF")+
# scale_fill_distiller(palette = "Blues")+
theme_minimal()+
scale_y_reverse(name = "Age (years)",lim= rev(c(0,6)),breaks = seq(0,6))+
scale_x_discrete(name = "Admission week",labels = leb_month)+
labs(tag = "D",fill = "Density")+
# theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1,size = 8))+
geom_line(data = ch,aes(x = date,y = c0,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c1,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c2,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c3,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c4,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c5,col = trend),
group = 1,lwd = 0.25)+
theme(axis.title.y = element_text(size = 18),
axis.title.x = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "top",
plot.tag = element_text(face = "bold", size = 18),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 18),
legend.text = element_text(size = 15),
legend.title = element_text(size = 18))+
guides(fill=guide_colourbar(barwidth=20,
label.position="top"),
color = "none")
hm2 <- ggplot(data=wwww$wdat, aes(x=date, y=age)) +
stat_density(
aes(fill = after_stat(count)),
geom = "raster",
position = "identity",
interpolate = TRUE
)+
scale_fill_paletteer_c("grDevices::Inferno")+
# scale_fill_gradient(low="#040404FF", high= "#FFFE9EFF")+
# scale_fill_distiller(palette = "Blues")+
theme_minimal()+
scale_y_reverse(name = "Age (years)",lim= rev(c(0,6)),breaks = seq(0,6))+
scale_x_discrete(name = "Admission week",labels = leb_month)+
labs(tag = "D",fill = "Number of\nhospitalizations")+
# theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1,size = 8))+
geom_line(data = ch,aes(x = date,y = c0,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c1,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c2,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c3,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c4,col = trend),
group = 1,lwd = 0.25)+
geom_line(data = ch,aes(x = date,y = c5,col = trend),
group = 1,lwd = 0.25)+
theme(axis.title.y = element_text(size = 18),
axis.title.x = element_blank(),
axis.ticks.x = element_blank(),
legend.position = "top",
plot.tag = element_text(face = "bold", size = 18),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 18),
legend.text = element_text(size = 15),
legend.title = element_text(size = 18))+
guides(fill=guide_colourbar(barwidth=20,
label.position="top"),
color = "none")
fi_peak <- df1 %>% filter(year(adm_date) == "2023") %>%
filter((adm_date <= as.Date("2023-09-03")&
!is.na(adm_date) & !is.na(age1)))
se_peak <- df1 %>% filter(year(adm_date) == "2023") %>%
filter((adm_date > as.Date("2023-09-03")) &
!is.na(adm_date) & !is.na(age1))
data <- data.frame(
peak = c( rep("1st",nrow(data.frame(se_peak$age1))),
rep("2nd",nrow(data.frame(fi_peak$age1)))),
age = c( fi_peak$age1, se_peak$age1 )
)
## density plot
ad <- ggplot(data=data, aes(x=age, group=peak, fill=peak)) +
geom_density(alpha = 0.6) +
scale_fill_manual(values = c("#582C83FF","#FFC72CFF")) +
scale_x_reverse(limit = c(6,0),
breaks = seq(0,6,by=1),
minor_breaks = NULL)+
scale_y_continuous(minor_breaks = NULL,position = "right")+
coord_flip()+
theme_minimal()+
labs(x = "Age", y ="Density",tag = "F",fill = "Wave")+
theme(axis.title.y = element_blank(),
axis.ticks.x = element_blank(),
# legend.position = "top",
plot.tag = element_text(face = "bold", size = 18),
axis.title.x = element_text(size = 18),
axis.text.x = element_text(size = 18),
axis.text.y = element_blank(),
legend.text = element_text(size = 18),
legend.title = element_text(size = 18))
library(ISOweek)
count_dangky_week <- readRDS("D:/OUCRU/hfmd/hcm_birth_data.rds")
child <- count_dangky_week %>% filter(birth_year >= 2017) %>% group_by(birth_week, birth_year) %>%
summarise(n=sum(n),.groups = "drop") %>% arrange(birth_year)
colnames(child) <- c("week","year","birth")
## combine week 52 and 53
child$week <- ifelse(child$week == 53,52,child$week)
child <- child %>% group_by(year) %>%
mutate(newn = ifelse(week == 52, sum(birth[week==52]), birth)) %>%
{data.frame(.$week, .$year, .$newn )} %>% unique() %>%
magrittr::set_colnames(c("week","year","birth"))
child$week2 <- seq(1:length(child$week))
time <- data.frame()
for (i in 2017:2022){
range <- child$week[child$year == i]
if (length(range) == 52){
time_add <- seq.int(i + 7/365 ,(i+1) - 7/365,
length.out = length(range)) %>% data.frame()
} else {
time_add <- seq.int(i + 7/365 ,(i+1) - 7/365,
length.out = 52)[1:length(range)] %>% data.frame()
}
time <- rbind(time,time_add)
}
child[,5] <- time %>%
magrittr::set_colnames(c("time"))
## model fitting
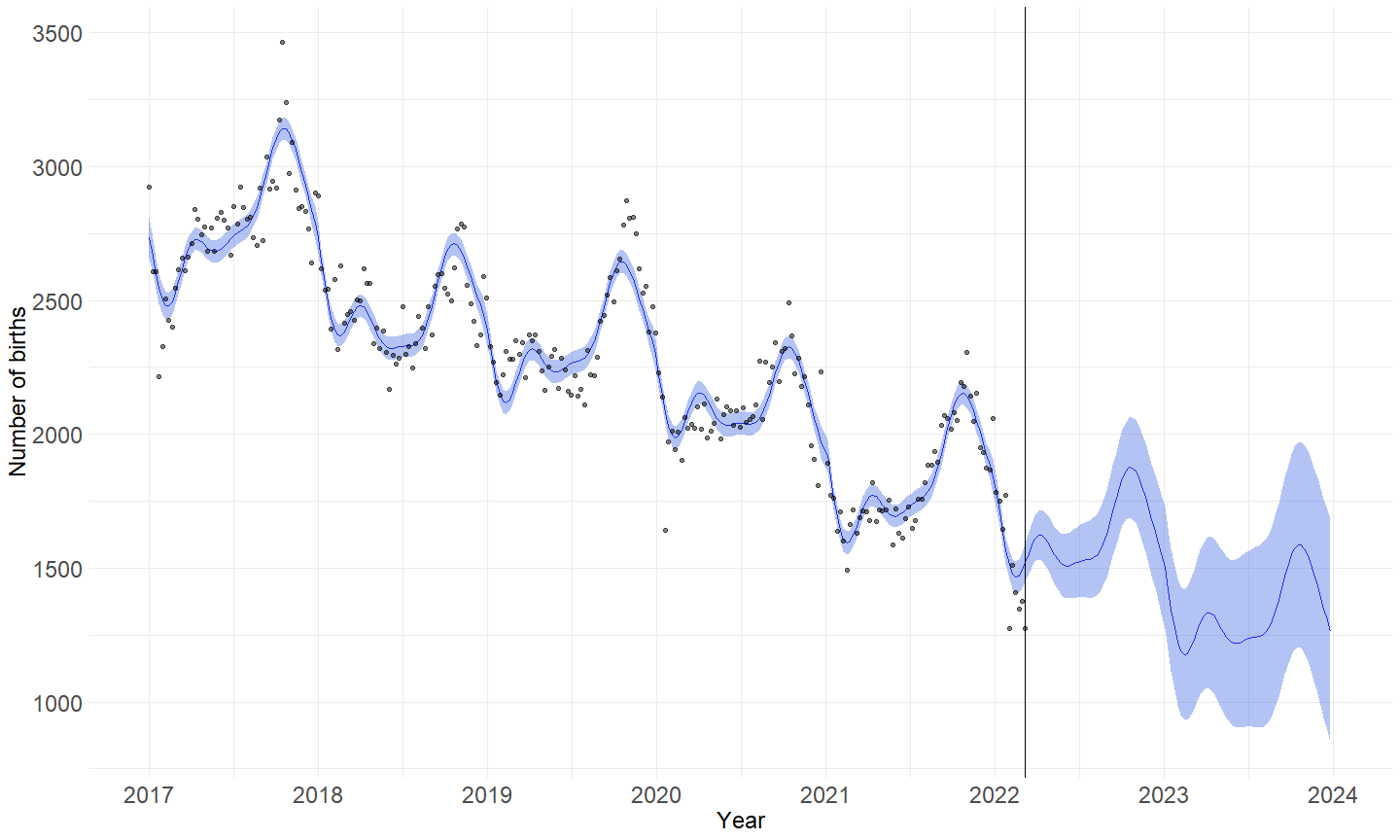
fit <- mgcv::gam(birth ~ s(week) + s(week2),method = "REML",data = child)
cutpoint <- function(point){
fitt <- mgcv::gam(birth ~ s(week) + s(week2),
method = "REML",data = child[-c(point:nrow(child)),])
new_data2 <- data.frame(week = rep(1:52,7))
new_data2$week2 <- seq(1,nrow(new_data2))
new_data2$year <- rep(2017:2023,each = 52)
time <- data.frame()
for (i in 2017:2023){
range <- new_data2$week[new_data2$year == i]
time_add <- data.frame(seq.int(i + 7/365 ,(i+1) - 7/365,
length.out = length(range)))
time <- rbind(time,time_add)
}
new_data2[4] <- time %>%
magrittr::set_colnames(c("time"))
est2 <- predict.gam(fitt,newdata = new_data2,
type="response",se.fit = TRUE)
new_data2 <- new_data2 %>% mutate(
fit = est2$fit,
lwr = est2$fit - qt(0.95,nrow(new_data2))*est2$se.fit,
upr = est2$fit + qt(0.95,nrow(new_data2))*est2$se.fit,
)
out <- list()
out$point <- point
out$df <- new_data2
return(out)
}
plot_cp <- function(model){
dta <- model$df %>%
mutate(time2 = sprintf("%04d-W%02d-1", year, week),
time2 = ISOweek2date(time2))
ggplot(data = dta) +
geom_line(aes(x = time2,y = fit),col = "blue")+
geom_ribbon(aes(x = time2,ymin = lwr, ymax = upr), fill = "royalblue",alpha = 0.4)+
geom_vline(xintercept = dta$time2[dta$week2 == model$point])+
labs(y = "Number of births",
x = "Year")+
theme_minimal()
}
model <- cutpoint(270)
b_ss <- plot_cp(model)+
geom_point(data = child[1:270,]%>%
mutate(time2 = sprintf("%04d-W%02d-1", year, week),
time2 = ISOweek2date(time2)) ,
aes(x = time2, y = birth),alpha = 0.5)+
# annotate("text", x= 2017, y=3500, label= "Fitting") +
# annotate("text", x = 2023.5, y=3500, label = "Estimation")+
scale_x_date(date_labels = "%Y",
breaks = seq(as.Date("2017-01-01"),
as.Date("2024-01-01"),
by = "1 year"),
limits = c(as.Date("2017-01-01"),
as.Date("2024-01-01")))+
theme(axis.title.y = element_text(size = 18),
axis.ticks.x = element_blank(),
legend.position = "bottom",
plot.tag = element_text(face = "bold", size = 18),
axis.title.x = element_text(size = 18),
axis.text.x = element_text(size = 18),
axis.text.y = element_text(size = 18),
legend.text = element_text(size = 15),
legend.title = element_text(size = 18))