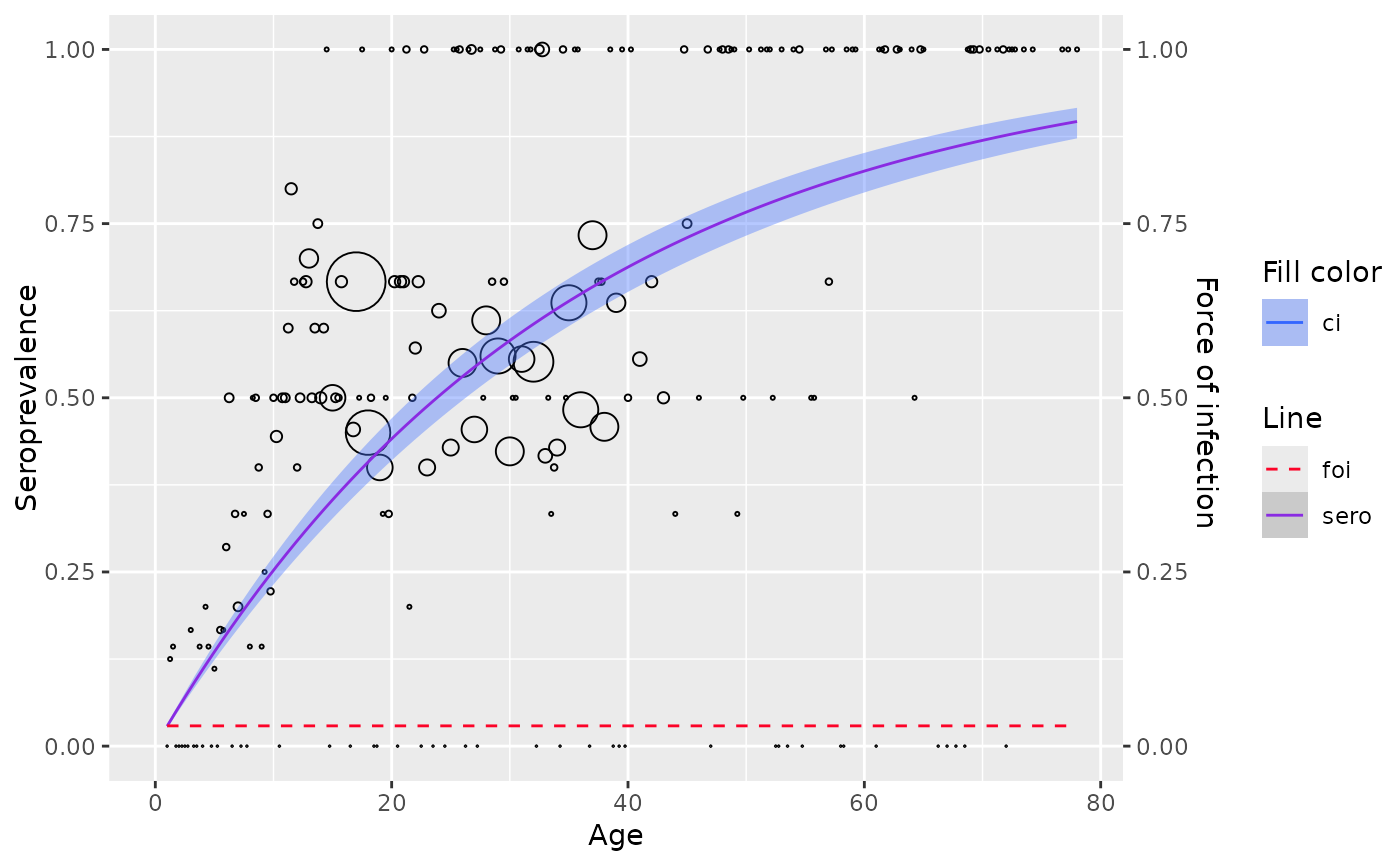
Refers to section 6.1.1
Arguments
- data
the input data frame, must either have `age`, `pos`, `tot` columns (for aggregated data) OR `age`, `status` for (linelisting data)
- k
degree of the model.
- type
name of method (Muench, Giffith, Grenfell).
- link
link function.
Value
a list of class polynomial_model with 5 items
- datatype
type of datatype used for model fitting (aggregated or linelisting)
- df
the dataframe used for fitting the model
- info
fitted "glm" object
- sp
seroprevalence
- foi
force of infection
Examples
data <- parvob19_fi_1997_1998[order(parvob19_fi_1997_1998$age), ]
data$status <- data$seropositive
aggregated <- transform_data(data$age, data$seropositive, stratum_col = "age")
# fit with aggregated data
model <- polynomial_model(aggregated, type = "Muench")
# fit with linelisting data
model <- polynomial_model(data, type = "Muench")
plot(model)