library(serosv)
#> Warning: replacing previous import 'magrittr::extract' by 'tidyr::extract' when
#> loading 'serosv'Visualize model
To visualize the model, user can simply use function
plot
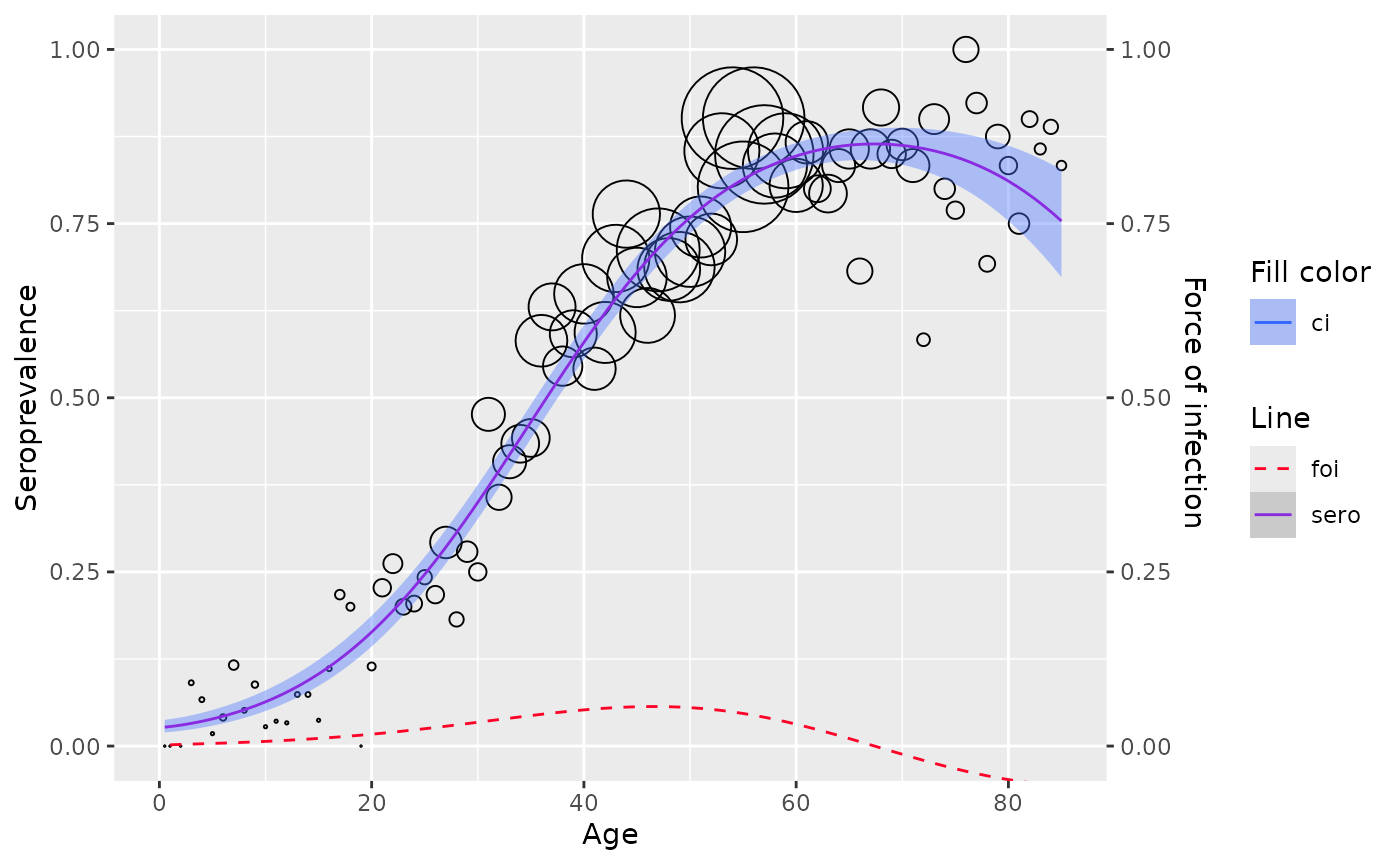
If implementation for confidence interval calculation is not yet
available, plot function would simply visualize
seroprevalence line
rubella <- rubella_uk_1986_1987
farrington_md <- farrington_model(
rubella,
start=list(alpha=0.07,beta=0.1,gamma=0.03)
)
plot(farrington_md)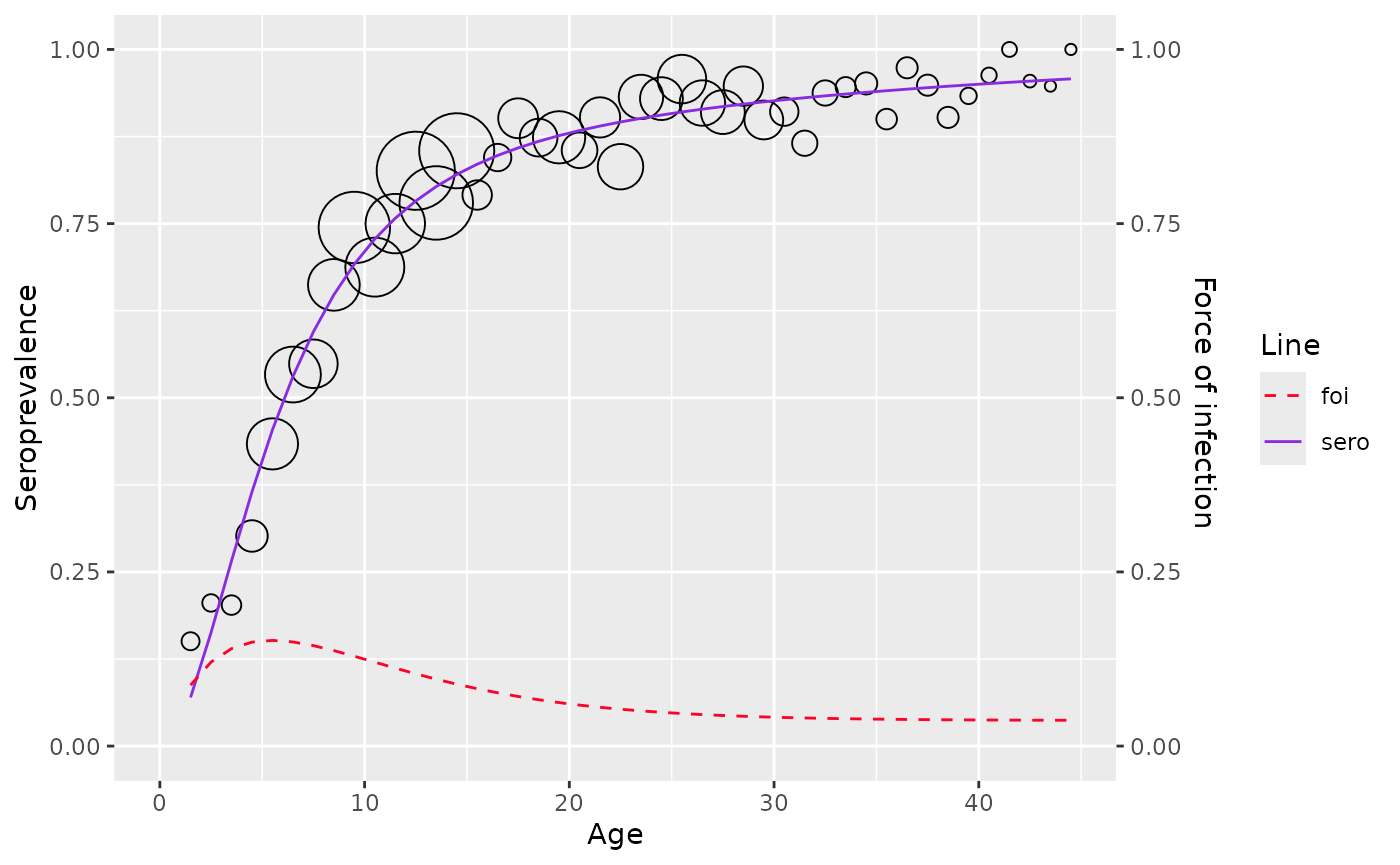
Customize the plot
Function set_plot_style() is provided to customize some
attributes of the plot.
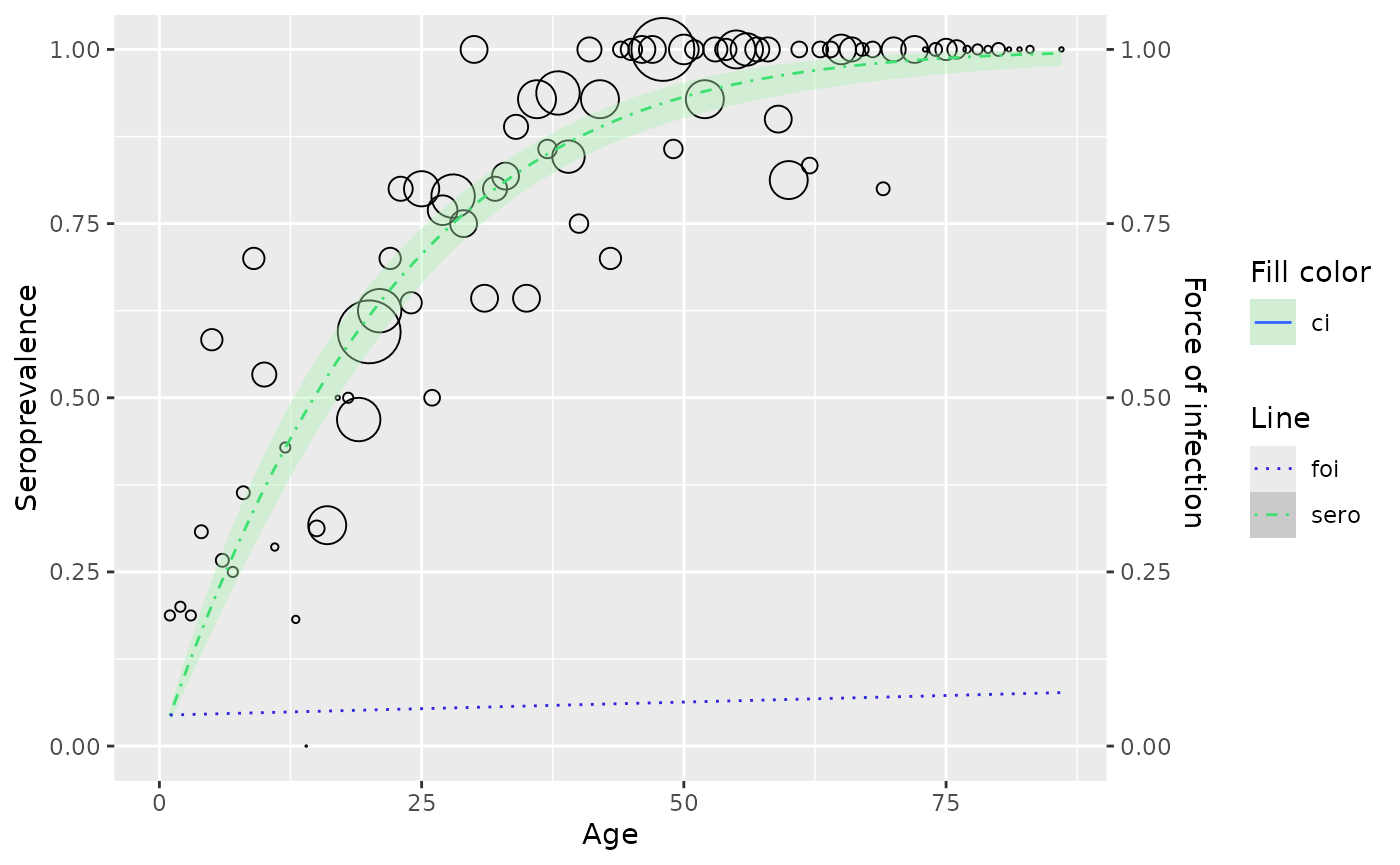
Current modifiable attributes include color, linetype for seroprevalence, foi and fill color for confidence interval
gf_model <- polynomial_model(hav_bg_1964, type = "Griffith")
# customize plot
plot(gf_model) +
set_plot_style(
sero = "#3de071",
foi = "#2f22e0",
ci = "#aaf2b2",
foi_line = "dotted",
sero_line = "dotdash"
)
#> Scale for colour is already present.
#> Adding another scale for colour, which will replace the existing scale.
#> Scale for linetype is already present.
#> Adding another scale for linetype, which will replace the existing scale.
#> Scale for fill is already present.
#> Adding another scale for fill, which will replace the existing scale.