Model repeated cross-sectional data
Source:vignettes/repeated_cross_sectional.Rmd
repeated_cross_sectional.RmdAge-time varying model
To monitor changes in a population’s seroprevalence over time, modelers often conduct multiple cross-sectional surveys at different time points, each using a new representative sample. The resulting data are known as repeated cross-sectional data.
Proposed approach
To model repeated cross-sectional serological data,
serosv offers age_time_model() function which
implements the following workflow:
- Fit age-specific seroprevalence for each survey period
- Monotonize age-specific or birth-cohort-specific prevalence over time
- Fit monotonized age-specific seroprevalence for each survey period
Fitting data
The function expects input data with the following columns:
Either
ageandstatus(linelisting) orageandpos+tot(aggregated)A column for date of survey (specified via
time_colargument)A column for id of each survey period (specified via
grouping_colargument)
# Prepare data
tb_nl <- tb_nl_1966_1973 %>%
mutate(
survey_year = age + birthyr,
survey_time = as.Date(paste0(survey_year, "-01-01"))
) %>% select(-birthyr) %>%
filter(survey_year > 1966) %>%
group_by(age, survey_year, survey_time) %>%
summarize(pos = sum(pos), tot = sum(tot), .groups = "drop")The monotonization method can be specified via the
monotonize_method argument, serosv currently
supports 2 options:
Pooled adjacent violators average (
monotonize_method = "pava")Shape constrained additive model (
monotonize_method = "scam")
The users can also configure to monotonize either:
Age-specific seroprevalence over time (
age_correct = FALSE)Or birth cohort specific seroprevalence over time (
age_correct=TRUE)
out_pava <- tb_nl %>%
age_time_model(
time_col = "survey_time",
grouping_col = "survey_year",
age_correct = F,
monotonize_method = "pava"
) %>%
suppressWarnings()
out_scam <- tb_nl %>%
age_time_model(
time_col = "survey_time",
grouping_col = "survey_year",
age_correct = T,
monotonize_method = "scam"
) %>%
suppressWarnings()The output is a data.frame with dimension [number of
survey, 9], where each row corresponds to a single survey period. The
columns are:
column for id of survey period
df- input data.frame corresponding to that survey periodinfo- model for the seroprevalencemonotonized_info- model for the monotonized seroprevalencemonotonized_ci_mod- model for the monotonized confidence intervalsp- predicted seroprevalence of the given input datafoi- estimated force of infection fromspmonotonized_sp- predicted monotonized seroprevalence of the given input datamonotonized_foi- estimated force of infection frommonotonized_sp
out_pava$out
#> # A tibble: 7 × 9
#> survey_year monotonized_info monotonized_ci_mod df info sp
#> <dbl> <list> <list> <list> <list> <list>
#> 1 1967 <gam> <named list [2]> <tibble> <gam> <dbl [5]>
#> 2 1968 <gam> <named list [2]> <tibble> <gam> <dbl [5]>
#> 3 1969 <gam> <named list [2]> <tibble> <gam> <dbl [6]>
#> 4 1970 <gam> <named list [2]> <tibble> <gam> <dbl [13]>
#> 5 1971 <gam> <named list [2]> <tibble> <gam> <dbl [8]>
#> 6 1972 <gam> <named list [2]> <tibble> <gam> <dbl [8]>
#> 7 1973 <gam> <named list [2]> <tibble> <gam> <dbl [5]>
#> # ℹ 3 more variables: foi <list>, monotonized_sp <list>, monotonized_foi <list>For visualization, the plot function for age_time_model
offers the following configurations
facetwhether to visualize result for each survey period separately (facet=TRUE) or on a single plot (facet=FALSE)modtypechoose which model to visualize, the model fitted with input data (modtype="non-monotonized") or monotonized data (modtype="monotonized")
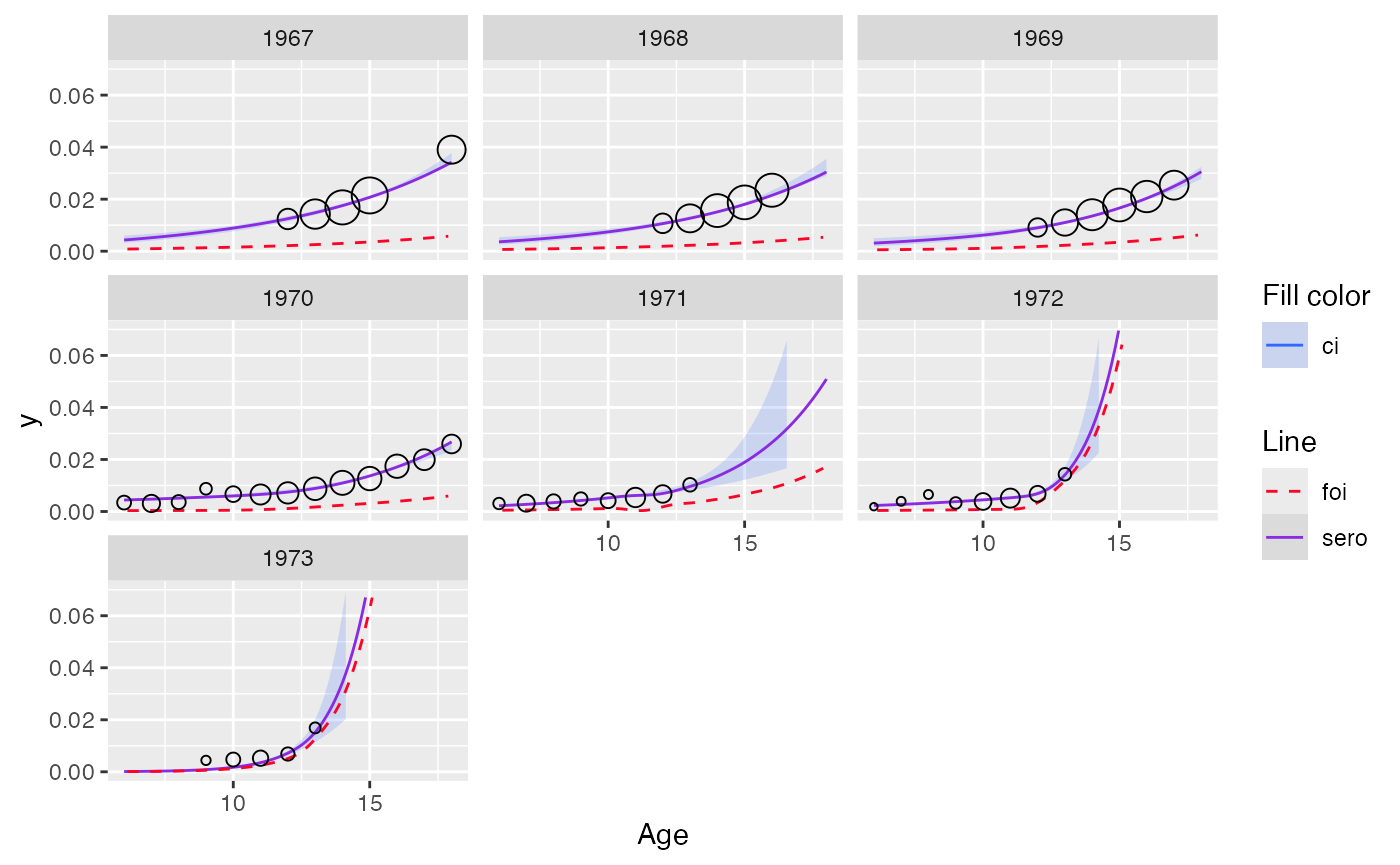
Example: output for model with PAVA for monotonization
plot(out_pava, facet = TRUE, modtype = "non-monotonized") + ylim(c(0, 0.07))
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.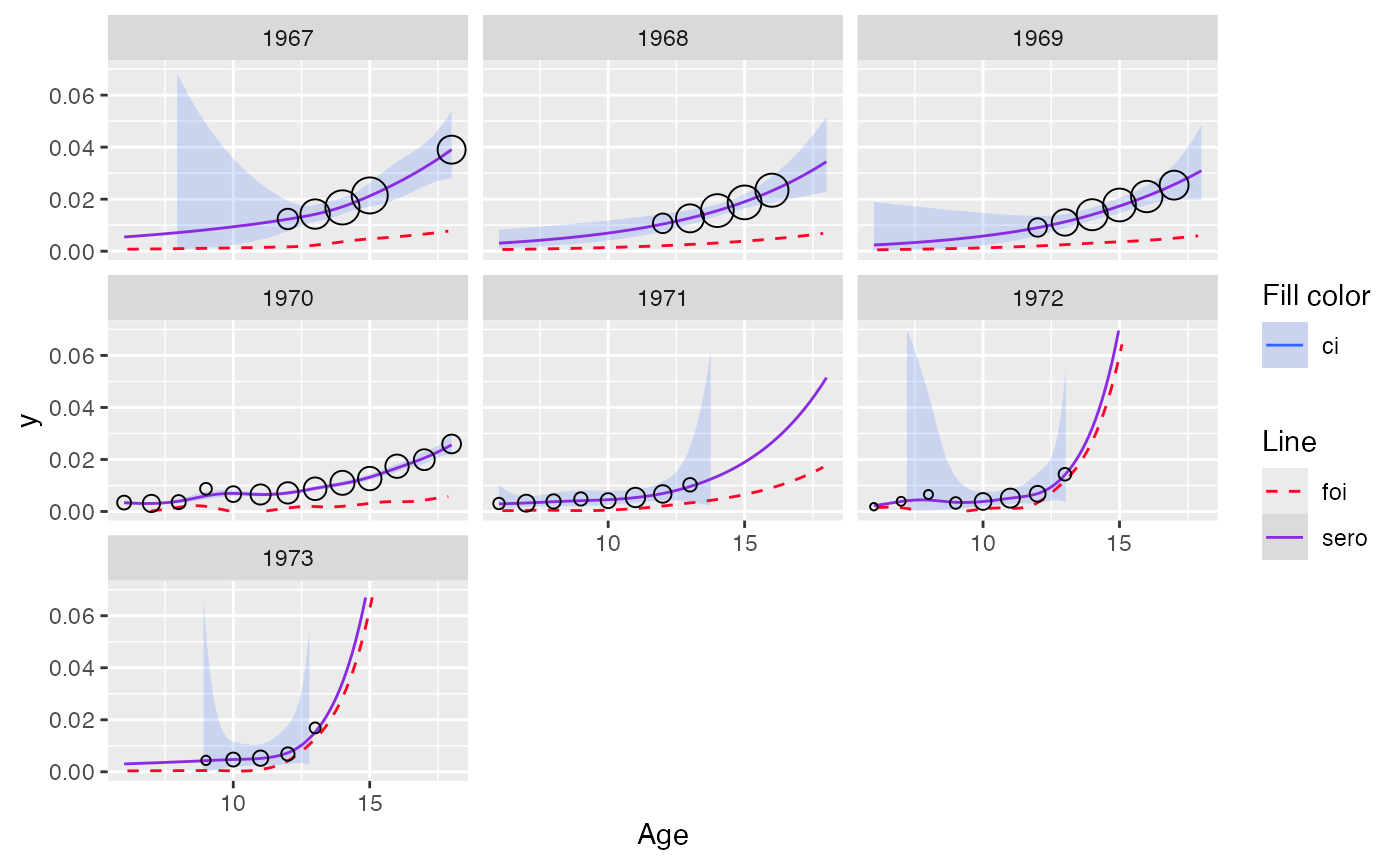
plot(out_pava, facet = TRUE, modtype = "monotonized") + ylim(c(0, 0.07))
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.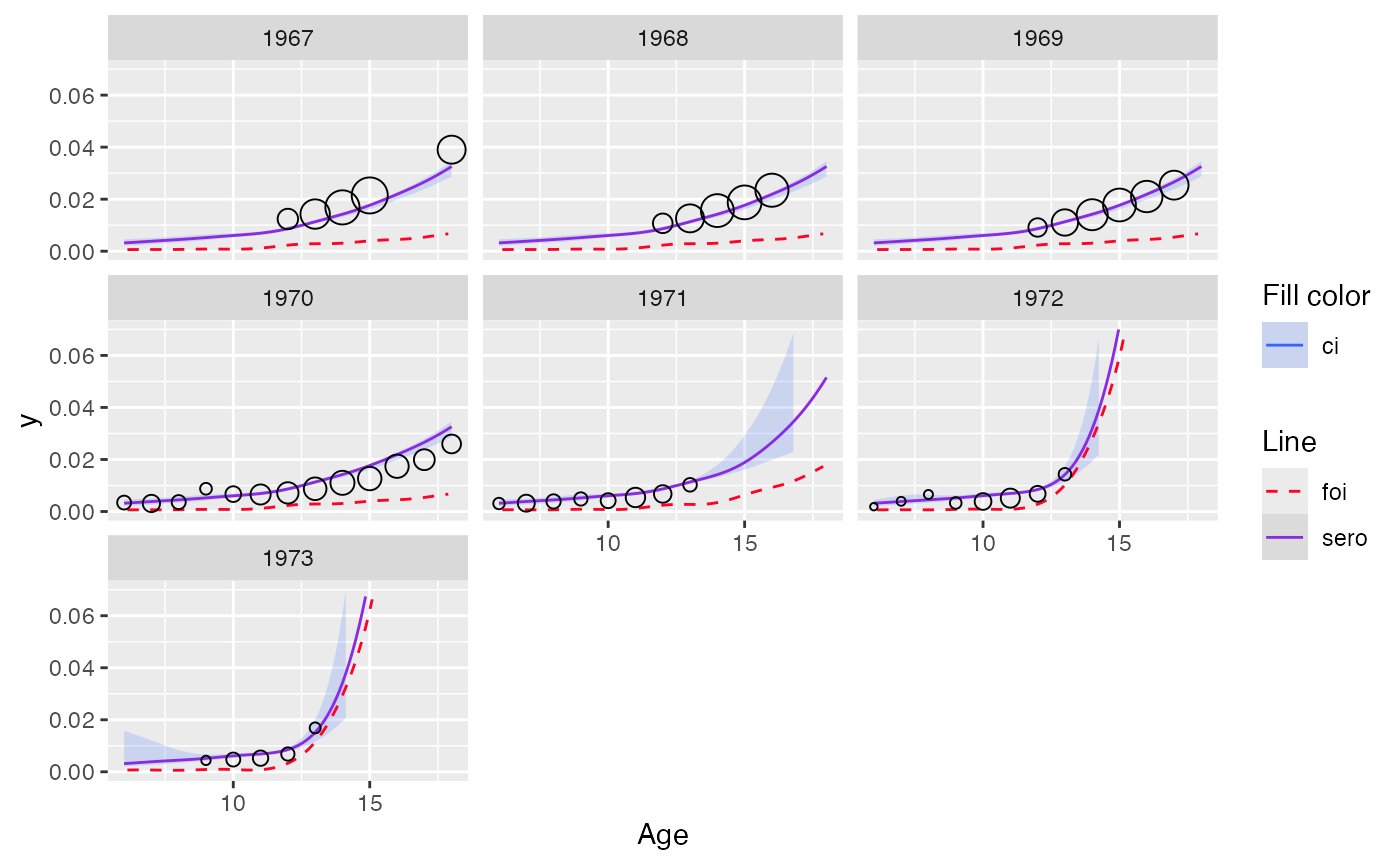
plot(out_pava, facet = FALSE, modtype = "monotonized") + ylim(c(0, 0.07))
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.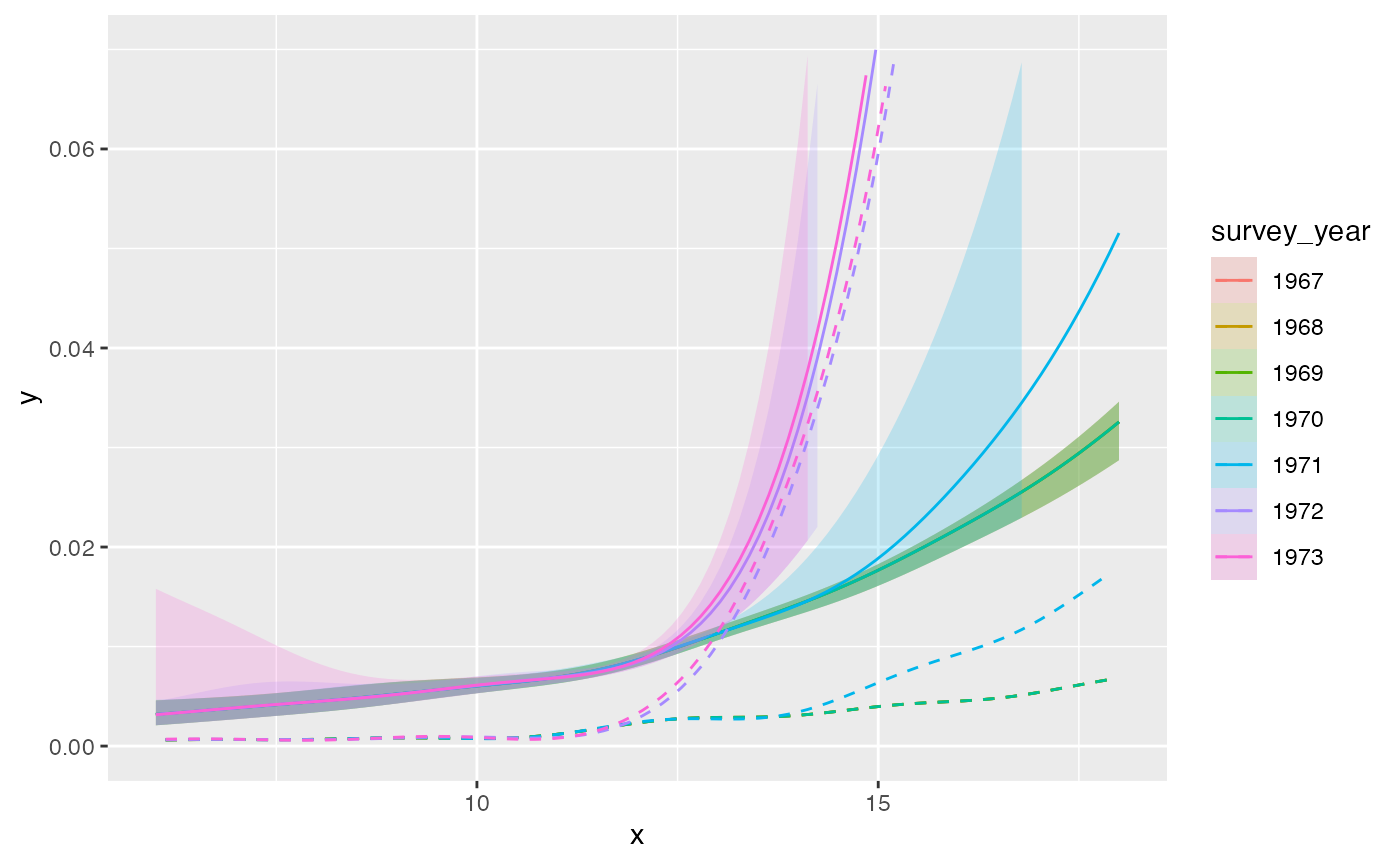
Example: output for model with SCAM for monotonization
plot(out_scam, facet = TRUE, modtype = "non-monotonized") + ylim(c(0, 0.07))
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.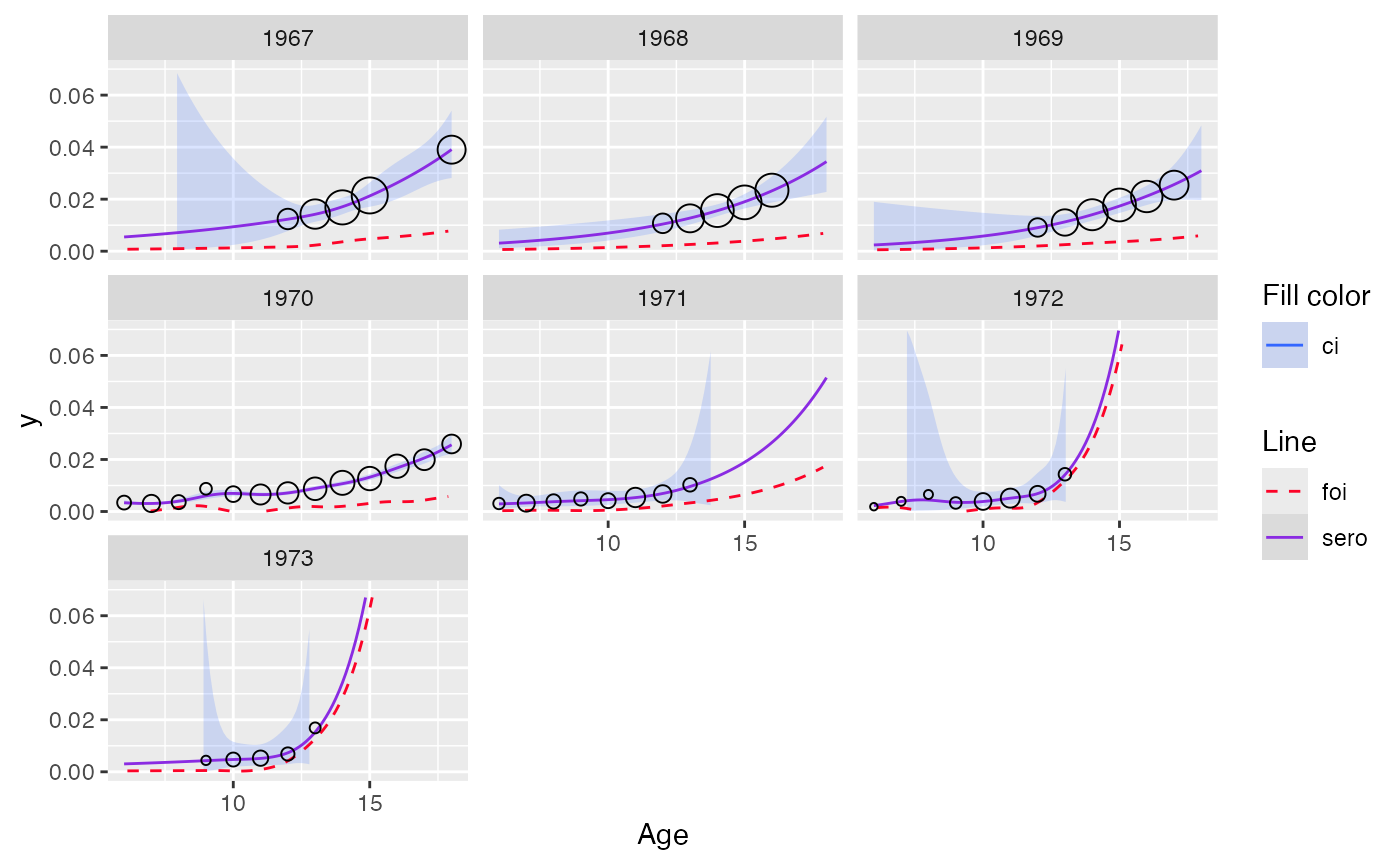
plot(out_scam, facet = TRUE, modtype = "monotonized") + ylim(c(0, 0.07))
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.